4GUY
 
 | | Human MMP12 catalytic domain in complex with*N*-Hydroxy-2-(2-(4-methoxyphenyl)ethylsulfonamido)acetamide | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-hydroxy-N~2~-{[2-(4-methoxyphenyl)ethyl]sulfonyl}glycinamide, ... | | Authors: | Calderone, V, Fragai, M, Luchinat, C, Massaro, A, Mordini, A, Mori, M. | | Deposit date: | 2012-08-30 | | Release date: | 2012-09-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contribution of free energy of solvation to ligand affinity in new potent MMPs inhibitors.
To be Published
|
|
2OY4
 
 | | Uninhibited human MMP-8 | | Descriptor: | CALCIUM ION, Neutrophil collagenase, ZINC ION | | Authors: | Calderone, V, Bertini, I, Fragai, M, Luchinat, C, Maletta, M, Yeo, K.J. | | Deposit date: | 2007-02-21 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Snapshots of the reaction mechanism of matrix metalloproteinases.
ANGEW.CHEM.INT.ED.ENGL., 45, 2006
|
|
2JQD
 
 | |
2OY2
 
 | | Human MMP-8 in complex with peptide IAG | | Descriptor: | CALCIUM ION, ILE-ALA-GLY peptide, Neutrophil collagenase, ... | | Authors: | Calderone, V, Bertini, I, Fragai, M, Luchinat, C, Maletta, M, Yeo, K.J. | | Deposit date: | 2007-02-21 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Snapshots of the reaction mechanism of matrix metalloproteinases.
ANGEW.CHEM.INT.ED.ENGL., 45, 2006
|
|
8VY6
 
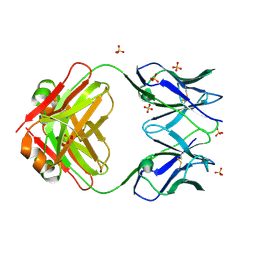 | | Murine light chain dimer | | Descriptor: | 6A8 light chain, SULFATE ION | | Authors: | Kapingidza, A.B, Dolamore, C, Hyduke, N.P, Easly, W, Chivv, C, Pomes, A, Chruszcz, M. | | Deposit date: | 2024-02-07 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural, Biophysical, and Computational Studies of a Murine Light Chain Dimer.
Molecules, 29, 2024
|
|
6YEL
 
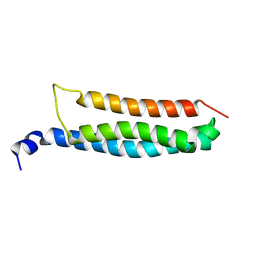 | | Stromal interaction molecule 1 coiled-coil 1 fragment | | Descriptor: | Stromal interaction molecule 1 | | Authors: | Rathner, P, Cerofolini, L, Ravera, E, Bechmann, M, Grabmayr, H, Fahrner, M, Fragai, M, Romanin, C, Luchinat, C, Mueller, N. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-02 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Interhelical interactions within the STIM1 CC1 domain modulate CRAC channel activation.
Nat.Chem.Biol., 17, 2021
|
|
5KW2
 
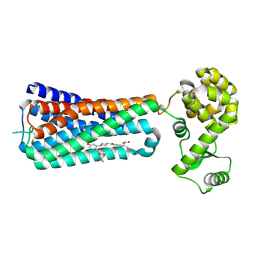 | | The extra-helical binding site of GPR40 and the structural basis for allosteric agonism and incretin stimulation | | Descriptor: | (3~{S})-3-cyclopropyl-3-[2-[1-[2-[2,2-dimethylpropyl-(6-methylpyridin-2-yl)carbamoyl]-5-methoxy-phenyl]piperidin-4-yl]-1-benzofuran-6-yl]propanoic acid, Free fatty acid receptor 1,Lysozyme,Free fatty acid receptor 1 | | Authors: | Ho, J.D, Chau, B, Rodgers, L, Lu, F, Wilbur, K.L, Otto, K.A, Chen, Y, Song, M, Riley, J.P, Yang, H.-C, Reynolds, N.A, Kahl, S.D, Lewis, A.P, Groshong, C, Madsen, R.E, Conners, K, Linswala, J.P, Gheyi, T, Saflor, M.D, Lee, M.R, Benach, J, Baker, K.A, Montrose-Rafizadeh, C, Genin, M.J, Miller, A.R, Hamdouchi, C. | | Deposit date: | 2016-07-15 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural basis for GPR40 allosteric agonism and incretin stimulation.
Nat Commun, 9, 2018
|
|
8XZ2
 
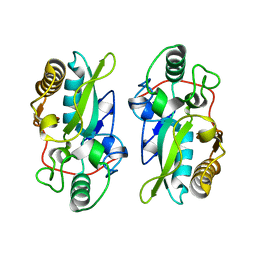 | | The structural model of a homodimeric D-Ala-D-Ala metallopeptidase, VanX, from vancomycin-resistant bacteria | | Descriptor: | D-alanyl-D-alanine dipeptidase | | Authors: | Konuma, T, Takai, T, Tsuchiya, C, Nishida, M, Hashiba, M, Yamada, Y, Shirai, H, Motoda, Y, Nagadoi, A, Chikaishi, E, Akagi, K, Akashi, S, Yamazaki, T, Akutsu, H, Oe, A, Ikegami, T. | | Deposit date: | 2024-01-20 | | Release date: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Analysis of the homodimeric structure of a D-Ala-D-Ala metallopeptidase, VanX, from vancomycin-resistant bacteria.
Protein Sci., 33, 2024
|
|
1CLF
 
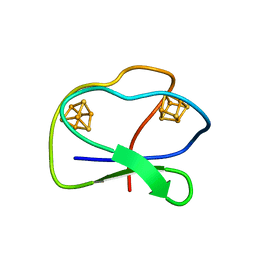 | | CLOSTRIDIUM PASTEURIANUM FERREDOXIN | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Bertini, I, Donaire, A, Feinberg, B.A, Luchinat, C, Piccioli, M, Yuan, H. | | Deposit date: | 1995-06-21 | | Release date: | 1996-01-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the oxidized 2[4Fe-4S] ferredoxin from Clostridium pasteurianum.
Eur.J.Biochem., 232, 1995
|
|
1OLR
 
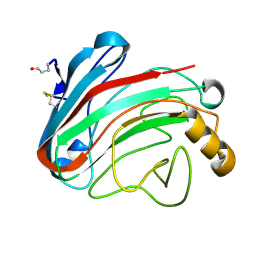 | | The Humicola grisea Cel12A Enzyme Structure at 1.2 A Resolution | | Descriptor: | ENDO-BETA-1,4-GLUCANASE | | Authors: | Sandgren, M, Gualfetti, P.J, Shaw, A, Gross, L.S, Saldajeno, M, Berglund, G.I, Jones, T.A, Mitchinson, C. | | Deposit date: | 2003-08-11 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Humicola Grisea Cel12A Enzyme Structure at 1.2 A Resolution and the Impact of its Free Cysteine Residues on Thermal Stability
Protein Sci., 12, 2003
|
|
1PYE
 
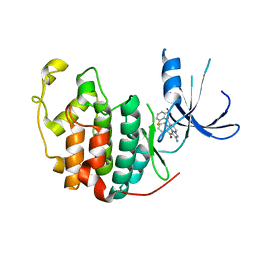 | | Crystal structure of CDK2 with inhibitor | | Descriptor: | Cell division protein kinase 2, [2-AMINO-6-(2,6-DIFLUORO-BENZOYL)-IMIDAZO[1,2-A]PYRIDIN-3-YL]-PHENYL-METHANONE | | Authors: | Zhang, F, Hamdouchi, C. | | Deposit date: | 2003-07-08 | | Release date: | 2004-07-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The discovery of a new structural class of cyclin-dependent kinase inhibitors, aminoimidazo[1,2-a]pyridines.
MOL.CANCER THER., 3, 2004
|
|
1OLQ
 
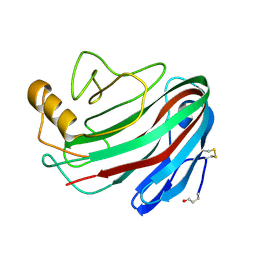 | | The Trichoderma reesei cel12a P201C mutant, structure at 1.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-BETA-1,4-GLUCANASE | | Authors: | Sandgren, M, Gualfetti, P.J, Shaw, A, Gross, L.S, Saldajeno, M, Berglund, G.I, Jones, T.A, Mitchinson, C. | | Deposit date: | 2003-08-11 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Humicola Grisea Cel12A Enzyme Structure at 1.2 A Resolution and the Impact of its Free Cysteine Residues on Thermal Stability
Protein Sci., 12, 2003
|
|
8TJF
 
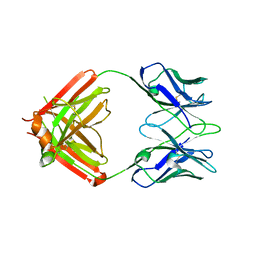 | | monovalent bispecific IgG antibodies through novel electrostatic steering mutations at the CH1-CL interface | | Descriptor: | Fab Lambda light chain, IgG1 Fab heavy chain | | Authors: | Oganesyan, V.Y, van Dyk, N, Mazor, Y, Chiang, C. | | Deposit date: | 2023-07-21 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Robust production of monovalent bispecific IgG antibodies through novel electrostatic steering mutations at the C H 1-C lambda interface.
Mabs, 15, 2023
|
|
1NOE
 
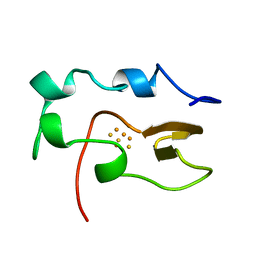 | | NMR STUDY OF REDUCED HIGH POTENTIAL IRON SULFUR PROTEIN | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Bentrop, D, Bertini, I, Capozzi, F, Dikiy, A, Eltis, L, Luchinat, C. | | Deposit date: | 1996-01-07 | | Release date: | 1996-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the reduced C77S mutant of the Chromatium vinosum high-potential iron-sulfur protein through nuclear magnetic resonance: comparison with the solution structure of the wild-type protein.
Biochemistry, 35, 1996
|
|
6TI5
 
 | | A New Structural Model of Abeta(1-40) Fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Bertini, I, Gonnelli, L, Luchinat, C, Mao, J, Nesi, A. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
1KSM
 
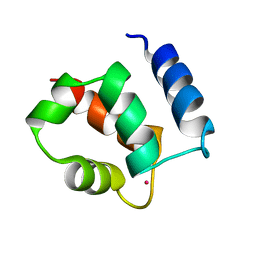 | | AVERAGE NMR SOLUTION STRUCTURE OF CA LN CALBINDIN D9K | | Descriptor: | LANTHANUM (III) ION, VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN | | Authors: | Bertini, I, Donaire, A, Luchinat, C, Piccioli, M, Poggi, L, Parigi, G, Jimenez, B. | | Deposit date: | 2002-01-14 | | Release date: | 2002-01-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Paramagnetism-based versus classical constraints: an analysis of the solution structure of Ca Ln calbindin D9k.
J.Biomol.NMR, 21, 2001
|
|
2HY9
 
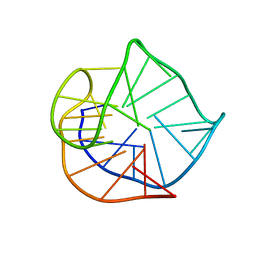 | | Human telomere DNA quadruplex structure in K+ solution hybrid-1 form | | Descriptor: | DNA (26-MER) | | Authors: | Dai, J, Punchihewa, C, Jones, R.A, Hurley, L, Yang, D. | | Deposit date: | 2006-08-04 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the intramolecular human telomeric G-quadruplex in potassium solution: a novel adenine triple formation.
Nucleic Acids Res., 35, 2007
|
|
1PFD
 
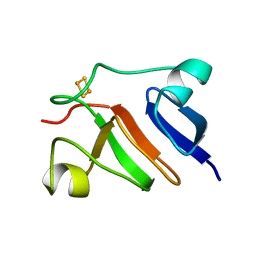 | | THE SOLUTION STRUCTURE OF HIGH PLANT PARSLEY [2FE-2S] FERREDOXIN, NMR, 18 STRUCTURES | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Im, S.-C, Liu, G, Luchinat, C, Sykes, A.G, Bertini, I. | | Deposit date: | 1998-05-05 | | Release date: | 1999-05-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of parsley [2Fe-2S]ferredoxin.
Eur.J.Biochem., 258, 1998
|
|
5J93
 
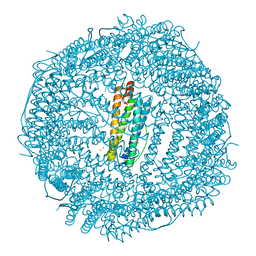 | | Five minutes iron loaded Rana Catesbeiana H' ferritin variant E57A/E136A/D140A | | Descriptor: | CHLORIDE ION, FE (II) ION, Ferritin, ... | | Authors: | Pozzi, C, Di Pisa, F, Mangani, S, Bernacchioni, C, Turano, P. | | Deposit date: | 2016-04-08 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ferroxidase Activity in Eukaryotic Ferritin is Controlled by Accessory-Iron-Binding Sites in the Catalytic Cavity.
Chemistry, 22, 2016
|
|
2K4G
 
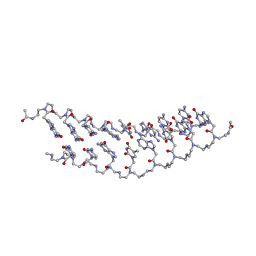 | | Solution Structure of a Peptide Nucleic Acid Duplex, 10 structures | | Descriptor: | METHYLAMINE, PNA (N'-(*(GPN)*(GPN)*(CPN)*(APN)*(TPN)*(GPN)*(CPN)*(CPN))-C') | | Authors: | He, W, Hatcher, E, Balaeff, A, Beratan, D, Gil, R, Madrid, M, Achim, C. | | Deposit date: | 2008-06-07 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a peptide nucleic acid duplex from NMR data: features and limitations.
J.Am.Chem.Soc., 130, 2008
|
|
6EOK
 
 | | Crystal structure of E. coli L-asparaginase II | | Descriptor: | L-asparaginase 2, ZINC ION | | Authors: | Cerofolini, L, Giuntini, S, Carlon, A, Ravera, E, Calderone, V, Fragai, M, Parigi, G, Luchinat, C. | | Deposit date: | 2017-10-09 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of PEGylated Asparaginase: New Opportunities from NMR Analysis of Large PEGylated Therapeutics.
Chemistry, 25, 2019
|
|
2GHJ
 
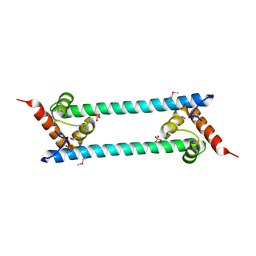 | | Crystal structure of folded and partially unfolded forms of Aquifex aeolicus ribosomal protein L20 | | Descriptor: | 50S ribosomal protein L20, SULFATE ION | | Authors: | Timsit, Y, Allemand, F, Chiaruttini, C, Springer, M. | | Deposit date: | 2006-03-27 | | Release date: | 2006-04-18 | | Last modified: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Coexistence of two protein folding states in the crystal structure of ribosomal protein L20
Embo Rep., 7, 2006
|
|
2JPZ
 
 | | Human telomere DNA quadruplex structure in K+ solution hybrid-2 form | | Descriptor: | DNA (26-MER) | | Authors: | Dai, J, Carver, M, Punchihewa, C, Jones, R, Yang, D. | | Deposit date: | 2007-05-25 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence
Nucleic Acids Res., 35, 2007
|
|
2JXY
 
 | | Solution structure of the hemopexin-like domain of MMP12 | | Descriptor: | CALCIUM ION, Macrophage metalloelastase | | Authors: | Bertini, I, Calderone, V, Fragai, M, Jaiswal, R, Luchinat, C, Melikian, M. | | Deposit date: | 2007-12-01 | | Release date: | 2008-05-27 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Evidence of reciprocal reorientation of the catalytic and hemopexin-like domains of full-length MMP-12
J.Am.Chem.Soc., 130, 2008
|
|
2K0J
 
 | | Solution structure of CaM complexed to DRP1p | | Descriptor: | CALCIUM ION, LANTHANUM (III) ION, calmodulin | | Authors: | Bertini, I, Luchinat, C, Parigi, G, Yuan, J, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2008-02-04 | | Release date: | 2009-03-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Accurate solution structures of proteins from X-ray data and a minimal set of NMR data: calmodulin-peptide complexes as examples.
J.Am.Chem.Soc., 131, 2009
|
|
