1T3Y
 
 | |
5D0N
 
 | | Crystal structure of maize PDRP bound with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Pyruvate, ... | | Authors: | Jiang, L, Chen, Z. | | Deposit date: | 2015-08-03 | | Release date: | 2016-02-24 | | Last modified: | 2016-03-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Reversible Phosphorylation by Maize Pyruvate Orthophosphate Dikinase Regulatory Protein
Plant Physiol., 170, 2016
|
|
5D1F
 
 | | Crystal structure of maize PDRP bound with AMP and Hg2+ | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, MERCURY (II) ION, ... | | Authors: | Jiang, L, Chen, Z. | | Deposit date: | 2015-08-04 | | Release date: | 2016-02-24 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Reversible Phosphorylation by Maize Pyruvate Orthophosphate Dikinase Regulatory Protein
Plant Physiol., 170, 2016
|
|
1TQF
 
 | | Crystal structure of human Beta secretase complexed with inhibitor | | Descriptor: | 3-{2-[(5-AMINOPENTYL)AMINO]-2-OXOETHOXY}-5-({[1-(4-FLUOROPHENYL)ETHYL]AMINO}CARBONYL)PHENYL PHENYLMETHANESULFONATE, Beta-secretase 1 | | Authors: | Munshi, S, Chen, Z, Kuo, L. | | Deposit date: | 2004-06-17 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a small molecule nonpeptide active site beta-secretase inhibitor that displays a nontraditional binding mode for aspartyl proteases.
J.Med.Chem., 47, 2004
|
|
3BV9
 
 | | Structure of Thrombin Bound to the Inhibitor FM19 | | Descriptor: | FM19 inhibitor, GLYCEROL, IODIDE ION, ... | | Authors: | Nieman, M.T, Burke, F, Warnock, M, Zhou, Y, Sweigert, J, Chen, A, Ricketts, D, Lucchesi, B.R, Chen, Z, Di Cera, E, Hilfinger, J, Mosberg, H.I, Schmaier, A.H. | | Deposit date: | 2008-01-05 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thrombostatin FM compounds: direct thrombin inhibitors - mechanism of action in vitro and in vivo.
J.Thromb.Haemost., 6, 2008
|
|
6TN8
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound BI-9564 | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 4-[4-[(dimethylamino)methyl]-2,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, ... | | Authors: | Williams, E.P, Chen, Z, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-12-06 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound BI-9564
To Be Published
|
|
7MIS
 
 | | Cryo-EM structure of SidJ-SdeC-CaM reaction intermediate complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Osinski, A, Black, M.H, Pawlowski, K, Chen, Z, Li, Y, Tagliabracci, V.S. | | Deposit date: | 2021-04-17 | | Release date: | 2021-08-18 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural and mechanistic basis for protein glutamylation by the kinase fold.
Mol.Cell, 81, 2021
|
|
7MIR
 
 | | Cryo-EM structure of SidJ-SdeA-CaM reaction intermediate complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Osinski, A, Black, M.H, Pawlowski, K, Chen, Z, Li, Y, Tagliabracci, V.S. | | Deposit date: | 2021-04-17 | | Release date: | 2021-08-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural and mechanistic basis for protein glutamylation by the kinase fold.
Mol.Cell, 81, 2021
|
|
7NNS
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound Momelotinib | | Descriptor: | 1,2-ETHANEDIOL, Activin receptor type I, Momelotinib, ... | | Authors: | Williams, E, Chen, Z, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound Momelotinib
To Be Published
|
|
6P9U
 
 | | Crystal structure of human thrombin mutant W215A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prothrombin, ZINC ION | | Authors: | Pelc, L.A, Koester, S.K, Chen, Z, Di Cera, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Residues W215, E217 and E192 control the allosteric E*-E equilibrium of thrombin.
Sci Rep, 9, 2019
|
|
1GG4
 
 | |
2PKA
 
 | | REFINED 2 ANGSTROMS X-RAY CRYSTAL STRUCTURE OF PORCINE PANCREATIC KALLIKREIN A, A SPECIFIC TRYPSIN-LIKE SERINE PROTEINASE. CRYSTALLIZATION, STRUCTURE DETERMINATION, CRYSTALLOGRAPHIC REFINEMENT, STRUCTURE AND ITS COMPARISON WITH BOVINE TRYPSIN | | Descriptor: | BENZAMIDINE, KALLIKREIN A | | Authors: | Bode, W, Chen, Z. | | Deposit date: | 1984-05-21 | | Release date: | 1984-07-19 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Refined 2 A X-ray crystal structure of porcine pancreatic kallikrein A, a specific trypsin-like serine proteinase. Crystallization, structure determination, crystallographic refinement, structure and its comparison with bovine trypsin.
J.Mol.Biol., 164, 1983
|
|
4FWJ
 
 | | Native structure of LSD2/AOF1/KDM1b in spacegroup of I222 at 2.9A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1B, PHOSPHATE ION, ... | | Authors: | Zhang, Q, Chen, Z. | | Deposit date: | 2012-07-01 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b
Cell Res., 23, 2013
|
|
2PGQ
 
 | | Human thrombin mutant C191A-C220A in complex with the inhibitor PPACK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, Thrombin heavy chain, ... | | Authors: | Bush-Pelc, L.A, Marino, F, Chen, Z, Pineda, A.O, Mathews, F.S, Di Cera, E. | | Deposit date: | 2007-04-10 | | Release date: | 2007-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Important role of the cys-191 cys-220 disulfide bond in thrombin function and allostery
J.Biol.Chem., 282, 2007
|
|
2PGB
 
 | | Inhibitor-free human thrombin mutant C191A-C220A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prothrombin, SULFATE ION | | Authors: | Bush-Pelc, L.A, Marino, F, Chen, Z, Pineda, A.O, Mathews, F.S, Di Cera, E. | | Deposit date: | 2007-04-09 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Important role of the cys-191 cys-220 disulfide bond in thrombin function and allostery
J.Biol.Chem., 282, 2007
|
|
3KLX
 
 | |
3KL1
 
 | |
6BJR
 
 | | Crystal structure of prothrombin mutant S101C/A470C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Prothrombin, ... | | Authors: | Chinnaraj, M, Chen, Z, Pelc, L, Grese, Z, Bystranowska, D, Di Cera, E, Pozzi, N. | | Deposit date: | 2017-11-06 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structure of prothrombin in the closed form reveals new details on the mechanism of activation.
Sci Rep, 8, 2018
|
|
6C2W
 
 | | Crystal structure of human prothrombin mutant S101C/A470C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Prothrombin, ... | | Authors: | Chinnaraj, M, Chen, Z, Pelc, L, Grese, Z, Bystranowska, D, Di Cera, E, Pozzi, N. | | Deposit date: | 2018-01-09 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (4.12 Å) | | Cite: | Structure of prothrombin in the closed form reveals new details on the mechanism of activation.
Sci Rep, 8, 2018
|
|
4XXB
 
 | | Crystal structure of human MDM2-RPL11 | | Descriptor: | 60S ribosomal protein L11, BETA-MERCAPTOETHANOL, E3 ubiquitin-protein ligase Mdm2, ... | | Authors: | Zheng, J, Chen, Z. | | Deposit date: | 2015-01-30 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human MDM2 complexed with RPL11 reveals the molecular basis of p53 activation
Genes Dev., 29, 2015
|
|
1INP
 
 | | CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 1-PHOSPHATASE AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | INOSITOL POLYPHOSPHATE 1-PHOSPHATASE, MAGNESIUM ION | | Authors: | York, J.D, Ponder, J.W, Chen, Z, Mathews, F.S, Majerus, P.W. | | Deposit date: | 1994-10-04 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of inositol polyphosphate 1-phosphatase at 2.3-A resolution.
Biochemistry, 33, 1994
|
|
4DT7
 
 | | Crystal structure of thrombin bound to the activation domain QEDQVDPRLIDGKMTRRGDS of protein C | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Pozzi, N, Barranco-Medina, S, Chen, Z, Di Cera, E. | | Deposit date: | 2012-02-20 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exposure of R169 controls protein C activation and autoactivation.
Blood, 120, 2012
|
|
7Q5U
 
 | | The tandem SH2 domains of SYK with a bound CD3G diphospho-ITAM peptide | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Chen, Z, Bountra, C, von Delft, F, Gileadi, O, Brennan, P.E. | | Deposit date: | 2021-11-04 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The tandem SH2 domains of SYK
To Be Published
|
|
7Q5T
 
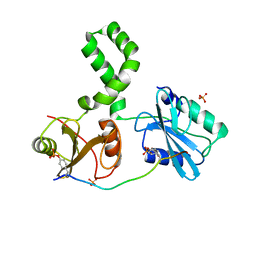 | | The tandem SH2 domains of SYK with a bound FCER1G diphospho-ITAM peptide | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, High affinity immunoglobulin epsilon receptor subunit gamma, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Chen, Z, Bountra, C, von Delft, F, Gileadi, O, Brennan, P.E. | | Deposit date: | 2021-11-04 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The tandem SH2 domains of SYK
To Be Published
|
|
7Q5W
 
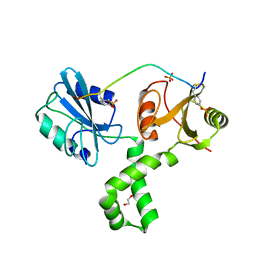 | | The tandem SH2 domains of SYK with a bound TYROBP diphospho-ITAM peptide | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Chen, Z, Bountra, C, von Delft, F, Gileadi, O, Brennan, P.E. | | Deposit date: | 2021-11-04 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The tandem SH2 domains of SYK
To Be Published
|
|
