6CKK
 
 | |
6BVH
 
 | | Trypsin complexed with a modified sunflower trypsin inhibitor, SFTI-TCTR(N12,N14) | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Riley, B.T, Chen, X. | | Deposit date: | 2017-12-13 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.927 Å) | | Cite: | Potent, multi-target serine protease inhibition achieved by a simplified beta-sheet motif.
PLoS ONE, 14, 2019
|
|
7JKS
 
 | | Crystal structure of vaccine-elicited broadly neutralizing VRC01-class antibody 2411a in complex with HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 gp120 core, The heavy chain of antibody 2411a, ... | | Authors: | Zhou, T, Chen, X, Kwong, P.D, Mascola, J.R. | | Deposit date: | 2020-07-28 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Vaccination induces maturation in a mouse model of diverse unmutated VRC01-class precursors to HIV-neutralizing antibodies with >50% breadth.
Immunity, 54, 2021
|
|
7JKT
 
 | | Crystal structure of vaccine-elicited broadly neutralizing VRC01-class antibody 2413a in complex with HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 gp120 core from strain d45-01dG5, ... | | Authors: | Zhou, T, Chen, X, Kwong, P.D, Mascola, J.R. | | Deposit date: | 2020-07-28 | | Release date: | 2021-06-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Vaccination induces maturation in a mouse model of diverse unmutated VRC01-class precursors to HIV-neutralizing antibodies with >50% breadth.
Immunity, 54, 2021
|
|
6CKM
 
 | |
6CKJ
 
 | |
6CU1
 
 | | X-ray structure of the S. typhimurium YrlA effector-binding module | | Descriptor: | MAGNESIUM ION, SULFATE ION, YrlA effector-binding module | | Authors: | Wang, W, Chen, X, Wolin, S.L, Xiong, Y. | | Deposit date: | 2018-03-23 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for tRNA Mimicry by a Bacterial Y RNA.
Structure, 26, 2018
|
|
6CKL
 
 | | N. meningitidis CMP-sialic acid synthetase in the presence of CMP and Neu5Ac2en | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Matthews, M.M, Fisher, A.J, Chen, X. | | Deposit date: | 2018-02-28 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.684 Å) | | Cite: | Catalytic Cycle ofNeisseria meningitidisCMP-Sialic Acid Synthetase Illustrated by High-Resolution Protein Crystallography.
Biochemistry, 2019
|
|
6IHI
 
 | | Crystal structure of RasADH 3B3/I91V from Ralstonia.sp in complex with NADPH and A6O | | Descriptor: | (2R,3S)-2-ethyl-2-[(2E)-2-(6-methoxy-3,4-dihydro-2H-naphthalen-1-ylidene)ethyl]-3-oxidanyl-cyclopentan-1-one, Alclohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, H.L, Chen, X, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2018-09-30 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Efficient reductive desymmetrization of bulky 1,3-cyclodiketones enabled by structure-guided directed evolution of a carbonyl reductase.
Nat Catal, 2, 2019
|
|
8IS2
 
 | |
8YMX
 
 | |
8YMW
 
 | |
6IHH
 
 | | Crystal structure of RasADH F12 from Ralstonia.sp in complex with NADPH and A6O | | Descriptor: | (2R,3S)-2-ethyl-2-[(2E)-2-(6-methoxy-3,4-dihydro-2H-naphthalen-1-ylidene)ethyl]-3-oxidanyl-cyclopentan-1-one, Alclohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, H.L, Chen, X, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2018-09-30 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Efficient reductive desymmetrization of bulky 1,3-cyclodiketones enabled by structure-guided directed evolution of a carbonyl reductase.
Nat Catal, 2, 2019
|
|
6KZQ
 
 | | structure of PTP-MEG2 and NSF-pY83 peptide complex | | Descriptor: | NSF-pY83 peptide, Tyrosine-protein phosphatase non-receptor type 9 | | Authors: | Xu, Y.F, Chen, X, Yu, X, Sun, J.P. | | Deposit date: | 2019-09-25 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | PTP-MEG2 regulates quantal size and fusion pore opening through two distinct structural bases and substrates.
Embo Rep., 22, 2021
|
|
6L03
 
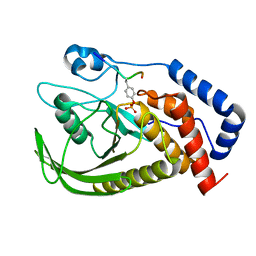 | | structure of PTP-MEG2 and MUNC18-1-pY145 peptide complex | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 9, stxbp1-pY145 peptide | | Authors: | Xu, Y.F, Chen, X, Yu, X, Sun, J.P. | | Deposit date: | 2019-09-25 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.084 Å) | | Cite: | PTP-MEG2 regulates quantal size and fusion pore opening through two distinct structural bases and substrates.
Embo Rep., 22, 2021
|
|
9BKX
 
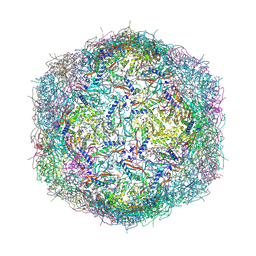 | | Mycobacterium tuberculosis encapsulin in complex with DyP | | Descriptor: | DODECAETHYLENE GLYCOL, Dye-decolorizing peroxidase, GLYCEROL, ... | | Authors: | Cuthbert, B.J, Batot, G.O, Contreras, H, Chen, X, Burley, K.H, Goulding, C.W. | | Deposit date: | 2024-04-29 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural Characterization of Mycobacterium tuberculosis Encapsulin in Complex with Dye-Decolorizing Peroxide.
Microorganisms, 12, 2024
|
|
6L6H
 
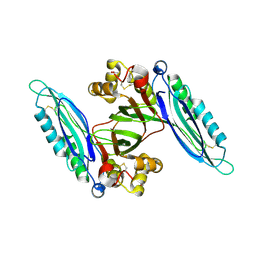 | | Crystal structure of Lpg0189 | | Descriptor: | ACETATE ION, GLYCEROL, Uncharacterized protein Lpg0189 | | Authors: | Ge, H, Chen, X. | | Deposit date: | 2019-10-29 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Crystal structure of a hypothetical T2SS effector Lpg0189 from Legionella pneumophila reveals a novel protein fold.
Biochem.Biophys.Res.Commun., 521, 2020
|
|
6L6G
 
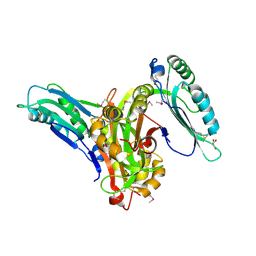 | | Crystal structure of SeMet_Lpg0189 | | Descriptor: | GLYCEROL, Uncharacterized protein Lpg0189 | | Authors: | Ge, H, Chen, X. | | Deposit date: | 2019-10-29 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of a hypothetical T2SS effector Lpg0189 from Legionella pneumophila reveals a novel protein fold.
Biochem.Biophys.Res.Commun., 521, 2020
|
|
5X7D
 
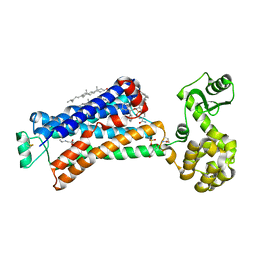 | | Structure of beta2 adrenoceptor bound to carazolol and an intracellular allosteric antagonist | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, 1,4-BUTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Liu, X, Ahn, S, Kahsai, A.W, Meng, K.-C, Latorraca, N.R, Pani, B, Venkatakrishnan, A.J, Masoudi, A, Weis, W.I, Dror, R.O, Chen, X, Lefkowitz, R.J, Kobilka, B.K. | | Deposit date: | 2017-02-25 | | Release date: | 2017-08-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Mechanism of intracellular allosteric beta 2AR antagonist revealed by X-ray crystal structure.
Nature, 548, 2017
|
|
8JQJ
 
 | | Crystal structure of carbonyl reductase SSCR mutant 1 from Sporobolomyces Salmonicolor | | Descriptor: | Aldehyde reductase 2 | | Authors: | Zhang, H.L, Li, Q, Liu, W.D, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineering a Carbonyl Reductase to Simultaneously Increase Activity Toward Bulky Ketone and Isopropanol for Dynamic Kinetic Asymmetric Reduction via Enzymatic Hydrogen Transfer
Acs Catalysis, 13, 2023
|
|
8JQK
 
 | | Crystal structure of a carbonyl reductase SSCR mutant from Sporobolomyces Salmonicolor | | Descriptor: | Aldehyde reductase 2 | | Authors: | Zhang, H.L, Li, Q, Liu, W.D, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Engineering a Carbonyl Reductase to Simultaneously Increase Activity Toward Bulky Ketone and Isopropanol for Dynamic Kinetic Asymmetric Reduction via Enzymatic Hydrogen Transfer
Acs Catalysis, 13, 2023
|
|
8H8Y
 
 | | Crystal structure of AbHheG from Acidimicrobiia bacterium | | Descriptor: | GLYCEROL, alpha/beta hydrolase | | Authors: | Zhou, C.H, Chen, X, Han, X, Liu, W.D, Wu, Q.Q, Zhu, D.M, Ma, Y.H. | | Deposit date: | 2022-10-24 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Flipping the Substrate Creates a Highly Selective Halohydrin Dehalogenase for the Synthesis of Chiral 4-Aryl-2-oxazolidinones from Readily Available Epoxides
Acs Catalysis, 13, 2023
|
|
7F5F
 
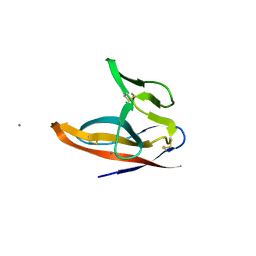 | | SARS-CoV-2 ORF8 S84 | | Descriptor: | CALCIUM ION, ORF8 protein | | Authors: | Chen, S, Zhou, Z, Chen, X. | | Deposit date: | 2021-06-22 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structures of Bat and Human Coronavirus ORF8 Protein Ig-Like Domain Provide Insights Into the Diversity of Immune Responses.
Front Immunol, 12, 2021
|
|
8HQP
 
 | | Crystal structure of AbHheG mutant from Acidimicrobiia bacterium | | Descriptor: | AbHheG_m | | Authors: | Zhou, C.H, Chen, X, Han, X, Liu, W.D, Wu, Q.Q, Zhu, D.M, Ma, Y.H. | | Deposit date: | 2022-12-13 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Flipping the Substrate Creates a Highly Selective Halohydrin Dehalogenase for the Synthesis of Chiral 4-Aryl-2-oxazolidinones from Readily Available Epoxides
Acs Catalysis, 13, 2023
|
|
5ZE2
 
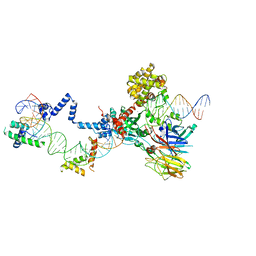 | | Hairpin Complex, RAG1/2-hairpin 12RSS/23RSS complex in 5mM Mn2+ for 2 min at 4'C | | Descriptor: | 1,2-ETHANEDIOL, DNA (30-MER), DNA (31-MER), ... | | Authors: | Kim, M.S, Chuenchor, W, Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2018-02-25 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination
Mol. Cell, 70, 2018
|
|
