6D1W
 
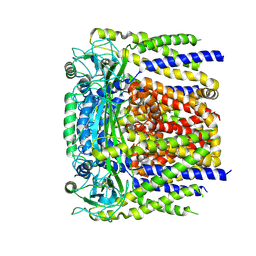 | | human PKD2 F604P mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Polycystin-2 | | Authors: | Zheng, W, Yang, X, Bulkley, D, Chen, X.Z, Cao, E. | | Deposit date: | 2018-04-12 | | Release date: | 2018-06-27 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Hydrophobic pore gates regulate ion permeation in polycystic kidney disease 2 and 2L1 channels.
Nat Commun, 9, 2018
|
|
8W2V
 
 | |
3VC8
 
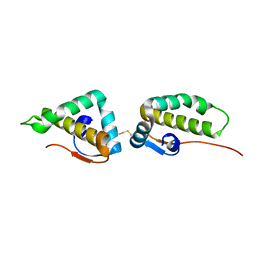 | | Crystal structure of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59 | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Xu, X, Lou, Z, Ma, Y, Chen, X, Yang, Z, Tong, X, Zhao, Q, Xu, Y, Deng, H, Bartlam, M, Rao, Z. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59.
Plos One, 4, 2009
|
|
3K6S
 
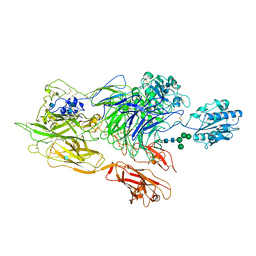 | | Structure of integrin alphaXbeta2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xie, C, Zhu, J, Chen, X, Mi, L, Nishida, N, Springer, T.A. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of an integrin with an alphaI domain, complement receptor type 4.
Embo J., 29, 2010
|
|
6IZJ
 
 | | Structural characterization of mutated NreA protein in nitrate binding site from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, NITRATE ION, NreA | | Authors: | Sangare, L, Chen, W, Wang, C, Chen, X, Wu, M, Zhang, X, Zang, J. | | Deposit date: | 2018-12-19 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the conformational change of Staphylococcus aureus NreA at C-terminus.
Biotechnol.Lett., 42, 2020
|
|
6KSE
 
 | |
6KPT
 
 | |
6KRI
 
 | |
3VV3
 
 | | Crystal structure of deseasin MCP-01 from Pseudoalteromonas sp. SM9913 | | Descriptor: | CALCIUM ION, Deseasin MCP-01 | | Authors: | Zhao, G.Y, Gao, X, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2012-07-14 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and mechanistic insights into collagen degradation by a bacterial collagenolytic serine protease in the subtilisin family.
Mol. Microbiol., 90, 2013
|
|
6K2H
 
 | | structural characterization of mutated NreA protein in nitrate binding site from staphylococcus aureus. | | Descriptor: | 1,2-ETHANEDIOL, NreA | | Authors: | Sangare, L, Chen, W, Wang, C, Chen, X, Wu, M, Zhang, X, Zang, J. | | Deposit date: | 2019-05-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the conformational change of Staphylococcus aureus NreA at C-terminus.
Biotechnol.Lett., 42, 2020
|
|
6KS9
 
 | |
6KSA
 
 | |
6KSB
 
 | |
2EX0
 
 | | Crystal structure of multifunctional sialyltransferase from Pasteurella Multocida | | Descriptor: | a2,3-sialyltransferase, a2,6-sialyltransferase | | Authors: | Ni, L, Sun, M, Chen, X, Fisher, A.J. | | Deposit date: | 2005-11-07 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cytidine 5'-Monophosphate (CMP)-Induced Structural Changes in a Multifunctional Sialyltransferase from Pasteurella multocida
Biochemistry, 45, 2006
|
|
3K71
 
 | | Structure of integrin alphaX beta2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xie, C, Zhu, J, Chen, X, Mi, L, Nishida, N, Springer, T.A. | | Deposit date: | 2009-10-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Structure of an integrin with an alphaI domain, complement receptor type 4.
Embo J., 29, 2010
|
|
7YGI
 
 | | Crystal structure of p53 DBD domain in complex with azurin | | Descriptor: | Azurin, Cellular tumor antigen p53, PHOSPHATE ION, ... | | Authors: | Jiang, W.X, Zuo, J.Q, Hu, J.J, Chen, X.Q, Ma, L.X, Liu, Z, Xing, Q. | | Deposit date: | 2022-07-11 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of bacterial effector protein azurin targeting tumor suppressor p53 and inhibiting its ubiquitination.
Commun Biol, 6, 2023
|
|
6IK4
 
 | | A Novel M23 Metalloprotease Pseudoalterin from Deep-sea | | Descriptor: | Elastinolytic metalloprotease, GLYCEROL, ZINC ION | | Authors: | Zhao, H.L, Tang, B.L, Yang, J, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2018-10-14 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A predator-prey interaction between a marine Pseudoalteromonas sp. and Gram-positive bacteria.
Nat Commun, 11, 2020
|
|
8W75
 
 | | Structure of Drosophila melanogaster L-2-hydroxyglutarate dehydrogenase | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FI05204p, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Yang, J, Chen, X, Jin, S, Ding, J. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and biochemical characterization of l-2-hydroxyglutarate dehydrogenase and its role in the pathogenesis of l-2-hydroxyglutaric aciduria.
J.Biol.Chem., 300, 2023
|
|
8W7F
 
 | | Structure of Drosophila melanogaster L-2-hydroxyglutarate dehydrogenase bound with FAD and a sulfate ion | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FI05204p, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yang, J, Chen, X, Jin, S, Ding, J. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structure and biochemical characterization of l-2-hydroxyglutarate dehydrogenase and its role in the pathogenesis of l-2-hydroxyglutaric aciduria.
J.Biol.Chem., 300, 2023
|
|
8W78
 
 | | Structure of Drosophila melanogaster L-2-hydroxyglutarate dehydrogenase in complex with FAD and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, DODECYL-BETA-D-MALTOSIDE, FI05204p, ... | | Authors: | Yang, J, Chen, X, Jin, S, Ding, J. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure and biochemical characterization of l-2-hydroxyglutarate dehydrogenase and its role in the pathogenesis of l-2-hydroxyglutaric aciduria.
J.Biol.Chem., 300, 2023
|
|
5X4G
 
 | | Crystal structure of Fab fragment of anti-CD147 monoclonal antibody 6H8 | | Descriptor: | 6H8 Fab heavy chain, 6H8 Fab light chain | | Authors: | Lin, P, Zhang, M.-Y, Chen, X, Ye, S, Yu, X.-L, Zhang, R.-G, Zhu, P, Chen, Z.-N. | | Deposit date: | 2017-02-13 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of CD147 C2 domain in complex with Fab of its monoclonal antibody
To Be Published
|
|
3K72
 
 | | Structure of integrin alphaX beta2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xie, C, Zhu, J, Chen, X, Mi, L, Nishida, N, Springer, T.A. | | Deposit date: | 2009-10-11 | | Release date: | 2010-01-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of an integrin with an alphaI domain, complement receptor type 4.
Embo J., 29, 2010
|
|
4R37
 
 | | Crystal structure analysis of LpxA, a UDP-N-acetylglucosamine acyltransferase from Bacteroides fragilis 9343 with UDP-GlcNAc | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETATE ION, ... | | Authors: | Fisher, A.J, Chen, X, Ngo, A. | | Deposit date: | 2014-08-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Bacteroides fragilis uridine 5'-diphosphate-N-acetylglucosamine (UDP-GlcNAc) acyltransferase (BfLpxA).
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7VF4
 
 | | Crystal structure of Vps75 from Candida albicans | | Descriptor: | CHLORIDE ION, SODIUM ION, Vps75 | | Authors: | Wang, W, Chen, X, Yang, Z, Chen, X, Li, C, Wang, M. | | Deposit date: | 2021-09-10 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of histone chaperone Vps75 from Candida albicans.
Biochem.Biophys.Res.Commun., 578, 2021
|
|
2H1L
 
 | |
