8JMP
 
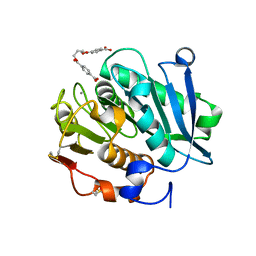 | | Structure of a leaf-branch compost cutinase, ICCG in complex with 1,4-butanediol terephthalate | | 分子名称: | 4-[4-(4-carboxyphenyl)carbonyloxybutoxycarbonyl]benzoic acid, CALCIUM ION, Leaf-branch compost cutinase | | 著者 | Yang, Y, Xue, T, Zheng, Y, Cheng, S, Guo, R.-T, Chen, C.-C. | | 登録日 | 2023-06-05 | | 公開日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Remodeling the polymer-binding cavity to improve the efficacy of PBAT-degrading enzyme.
J Hazard Mater, 464, 2023
|
|
8JMO
 
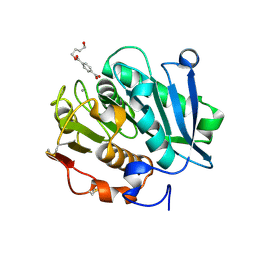 | | Structure of a leaf-branch compost cutinase, ICCG in complex with 4-((4-Hydroxybutoxy)carbonyl)benzoic acid | | 分子名称: | 4-(4-oxidanylbutoxycarbonyl)benzoic acid, CALCIUM ION, Leaf-branch compost cutinase | | 著者 | Yang, Y, Xue, T, Zheng, Y, Cheng, S, Guo, R.-T, Chen, C.-C. | | 登録日 | 2023-06-05 | | 公開日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Remodeling the polymer-binding cavity to improve the efficacy of PBAT-degrading enzyme.
J Hazard Mater, 464, 2023
|
|
6JYV
 
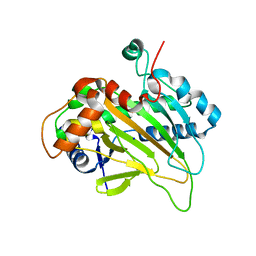 | | Structure of an isopenicillin N synthase from Pseudomonas aeruginosa PAO1 | | 分子名称: | Probable iron/ascorbate oxidoreductase, SODIUM ION | | 著者 | Hao, Z, Che, S, Wang, R, Liu, R, Zhang, Q, Bartlam, M. | | 登録日 | 2019-04-28 | | 公開日 | 2019-05-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.651 Å) | | 主引用文献 | Structural characterization of an isopenicillin N synthase family oxygenase from Pseudomonas aeruginosa PAO1.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
7WJL
 
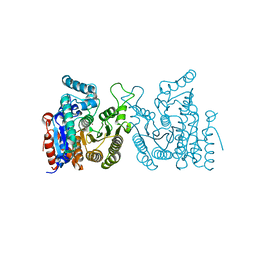 | | Crystal structure of S. cerevisiae Hos3 | | 分子名称: | ACETATE ION, Histone deacetylase HOS3, ZINC ION | | 著者 | Pang, N.N, Che, S.Y, Yang, N. | | 登録日 | 2022-01-07 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural characterization of fungus-specific histone deacetylase Hos3 provides insights into developing selective inhibitors with antifungal activity.
J.Biol.Chem., 298, 2022
|
|
6LFD
 
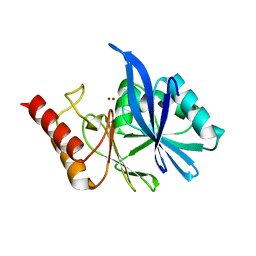 | |
8YUU
 
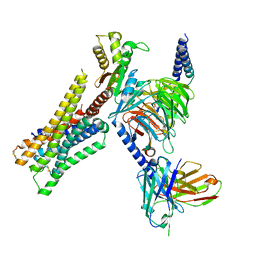 | | Cryo-EM structure of the histamine-bound H3R-Gi complex | | 分子名称: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Shen, Q, Tang, X, Wen, X, Cheng, S, Xiao, P, Zang, S, Shen, D, Jiang, L, Zheng, Y, Zhang, H, Xu, H, Mao, C, Zhang, M, Hu, W, Sun, J, Chen, Z, Zhang, Y. | | 登録日 | 2024-03-27 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Molecular Determinant Underlying Selective Coupling of Primary G-Protein by Class A GPCRs.
Adv Sci, 11, 2024
|
|
8YUV
 
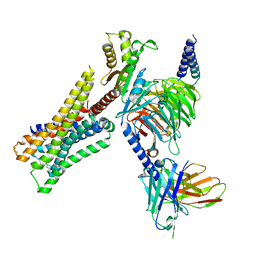 | | Cryo-EM structure of the immepip-bound H3R-Gi complex | | 分子名称: | 4-(1H-imidazol-5-ylmethyl)piperidine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Shen, Q, Tang, X, Wen, X, Cheng, S, Xiao, P, Zang, S, Shen, D, Jiang, L, Zheng, Y, Zhang, H, Xu, H, Mao, C, Zhang, M, Hu, W, Sun, J, Chen, Z, Zhang, Y. | | 登録日 | 2024-03-27 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Molecular Determinant Underlying Selective Coupling of Primary G-Protein by Class A GPCRs.
Adv Sci, 11, 2024
|
|
8YUT
 
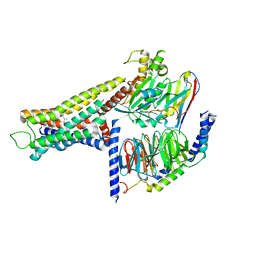 | | Cryo-EM structure of the amthamine-bound H2R-Gs complex | | 分子名称: | 5-(2-azanylethyl)-4-methyl-1,3-thiazol-2-amine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Shen, Q, Tang, X, Wen, X, Cheng, S, Xiao, P, Zang, S, Shen, D, Jiang, L, Zheng, Y, Zhang, H, Xu, H, Mao, C, Zhang, M, Hu, W, Sun, J, Chen, Z, Zhang, Y. | | 登録日 | 2024-03-27 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Molecular Determinant Underlying Selective Coupling of Primary G-Protein by Class A GPCRs.
Adv Sci, 11, 2024
|
|
6LF4
 
 | | Crystal structure of VMB-1 bound to hydrolyzed meropenem | | 分子名称: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, VMB-1 metallo-beta-lactamase, ZINC ION | | 著者 | Cheng, Q, Chen, S. | | 登録日 | 2019-11-28 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Crystal structure of VMB-1 bound to hydrolyzed meropenem
To Be Published
|
|
7Y72
 
 | | SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 (focused refinement on Fab-RBD interface) | | 分子名称: | Fab E7 heavy chain, Fab E7 light chain, Spike glycoprotein | | 著者 | Chia, W.N, Tan, C.W, Tan, A.W.K, Young, B, Starr, T.N, Lopez, E, Fibriansah, G, Barr, J, Cheng, S, Yeoh, A.Y.Y, Yap, W.C, Lim, B.L, Ng, T.S, Sia, W.R, Zhu, F, Chen, S, Zhang, J, Greaney, A.J, Chen, M, Au, G.G, Paradkar, P, Peiris, M, Chung, A.W, Bloom, J.D, Lye, D, Lok, S.M, Wang, L.F. | | 登録日 | 2022-06-21 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (4.03 Å) | | 主引用文献 | Potent pan huACE2-dependent sarbecovirus neutralizing monoclonal antibodies isolated from a BNT162b2-vaccinated SARS survivor.
Sci Adv, 9, 2023
|
|
7Y71
 
 | | SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab E7 heavy chain, ... | | 著者 | Chia, W.N, Tan, C.W, Tan, A.W.K, Young, B, Starr, T.N, Lopez, E, Fibriansah, G, Barr, J, Cheng, S, Yeoh, A.Y.Y, Yap, W.C, Lim, B.L, Ng, T.S, Sia, W.R, Zhu, F, Chen, S, Zhang, J, Greaney, A.J, Chen, M, Au, G.G, Paradkar, P, Peiris, M, Chung, A.W, Bloom, J.D, Lye, D, Lok, S.M, Wang, L.F. | | 登録日 | 2022-06-21 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.12 Å) | | 主引用文献 | Potent pan huACE2-dependent sarbecovirus neutralizing monoclonal antibodies isolated from a BNT162b2-vaccinated SARS survivor.
Sci Adv, 9, 2023
|
|
3OTW
 
 | | Structural and Functional Studies of Helicobacter pylori Wild-Type and Mutated Proteins Phosphopantetheine adenylyltransferase | | 分子名称: | COENZYME A, Phosphopantetheine adenylyltransferase, SULFATE ION | | 著者 | Yin, H.S, Cheng, C.S, Chen, C.G, Luo, Y.C, Chen, W.T, Cheng, S.Y. | | 登録日 | 2010-09-14 | | 公開日 | 2011-09-14 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural and Functional Studies of Helicobacter pylori Wild-Type and Mutated Proteins Phosphopantetheine adenylyltransferase
To be Published
|
|
6IMQ
 
 | | Crystal structure of PML B1-box multimers | | 分子名称: | CHLORIDE ION, Protein PML, ZINC ION | | 著者 | Li, Y, Ma, X, Chen, Z, Wu, H, Wang, P, Wu, W, Cheng, N, Zeng, L, Zhang, H, Cai, X, Chen, S.J, Chen, Z, Meng, G. | | 登録日 | 2018-10-23 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | B1 oligomerization regulates PML nuclear body biogenesis and leukemogenesis.
Nat Commun, 10, 2019
|
|
4XEH
 
 | |
4XDY
 
 | | Structure of NADH-preferring ketol-acid reductoisomerase from an uncultured archean | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, Ketol-acid reductoisomerase, ... | | 著者 | Cahn, J.K.B, Brinkmann-Chen, S, Arnold, F.H. | | 登録日 | 2014-12-20 | | 公開日 | 2015-04-22 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.535 Å) | | 主引用文献 | Cofactor specificity motifs and the induced fit mechanism in class I ketol-acid reductoisomerases.
Biochem.J., 468, 2015
|
|
8QN7
 
 | | Amyloid-beta 40 type 1 filament from the leptomeninges of individual with Alzheimer's disease and cerebral amyloid angiopathy | | 分子名称: | Amyloid-beta A4 protein | | 著者 | Yang, Y, Murzin, A.S, Peak-Chew, S.Y, Franco, C, Newell, K.L, Ghetti, B, Goedert, M, Scheres, S.H.W. | | 登録日 | 2023-09-25 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Cryo-EM structures of A beta 40 filaments from the leptomeninges of individuals with Alzheimer's disease and cerebral amyloid angiopathy.
Acta Neuropathol Commun, 11, 2023
|
|
8QN6
 
 | | Amyloid-beta 40 type 2 filament from the leptomeninges of individual with Alzheimer's disease and cerebral amyloid angiopathy | | 分子名称: | Amyloid-beta A4 protein | | 著者 | Yang, Y, Murzin, A.S, Peak-Chew, S.Y, Franco, C, Newell, K.L, Ghetti, B, Goedert, M, Scheres, S.H.W. | | 登録日 | 2023-09-25 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Cryo-EM structures of A beta 40 filaments from the leptomeninges of individuals with Alzheimer's disease and cerebral amyloid angiopathy.
Acta Neuropathol Commun, 11, 2023
|
|
4XDZ
 
 | | Holo structure of ketol-acid reductoisomerase from Ignisphaera aggregans | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Ketol-acid reductoisomerase, ... | | 著者 | Cahn, J.K.B, Brinkmann-Chen, S, Arnold, F.H. | | 登録日 | 2014-12-20 | | 公開日 | 2015-04-22 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Cofactor specificity motifs and the induced fit mechanism in class I ketol-acid reductoisomerases.
Biochem.J., 468, 2015
|
|
6J2O
 
 | | Crystal structure of CTX-M-64 clavulanic acid complex | | 分子名称: | (2E)-3-[(4-hydroxy-2-oxobutyl)amino]prop-2-enal, Beta-lactamase | | 著者 | Cheng, Q, Chen, S. | | 登録日 | 2019-01-02 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Insight into the Mechanism of Inhibitor Resistance in CTX-M-199, a CTX-M-64 Variant Carrying the S130T Substitution.
Acs Infect Dis., 6, 2020
|
|
6J25
 
 | | CTX-M-64 beta-lactamase mutant-S130T | | 分子名称: | Beta-lactamase | | 著者 | Cheng, Q, Chen, S. | | 登録日 | 2018-12-30 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structural Insight into the Mechanism of Inhibitor Resistance in CTX-M-199, a CTX-M-64 Variant Carrying the S130T Substitution.
Acs Infect Dis., 6, 2020
|
|
5ZB7
 
 | | CTX-M-64 apoenzyme | | 分子名称: | Beta-lactamase | | 著者 | Cheng, Q, Chen, S. | | 登録日 | 2018-02-10 | | 公開日 | 2019-02-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Structural Insight into the Mechanism of Inhibitor Resistance in CTX-M-199, a CTX-M-64 Variant Carrying the S130T Substitution.
Acs Infect Dis., 6, 2020
|
|
6J2K
 
 | | CTX-M-64 beta-lactamase S130T clavulanic acid complex | | 分子名称: | (2E)-3-[(4-hydroxy-2-oxobutyl)amino]prop-2-enal, Beta-lactamase | | 著者 | Cheng, Q, Chen, S. | | 登録日 | 2019-01-01 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Structural Insight into the Mechanism of Inhibitor Resistance in CTX-M-199, a CTX-M-64 Variant Carrying the S130T Substitution.
Acs Infect Dis., 6, 2020
|
|
6J2B
 
 | | CTX-M-64 beta-lactamase S130T sulbactam complex | | 分子名称: | Beta-lactamase, GLYCEROL, TRANS-ENAMINE INTERMEDIATE OF SULBACTAM | | 著者 | Cheng, Q, Chen, S. | | 登録日 | 2018-12-31 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Structural Insight into the Mechanism of Inhibitor Resistance in CTX-M-199, a CTX-M-64 Variant Carrying the S130T Substitution.
Acs Infect Dis., 6, 2020
|
|
4TSK
 
 | | Ketol-acid reductoisomerase from Alicyclobacillus acidocaldarius | | 分子名称: | Ketol-acid reductoisomerase, L(+)-TARTARIC ACID, MAGNESIUM ION, ... | | 著者 | Cahn, J.K.B, Brinkmann-Chen, S, Arnold, F.H. | | 登録日 | 2014-06-18 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Uncovering rare NADH-preferring ketol-acid reductoisomerases.
Metab. Eng., 26C, 2014
|
|
6JV4
 
 | | Crystal structure of metallo-beta-lactamase VMB-1 | | 分子名称: | CITRIC ACID, VMB-1, ZINC ION | | 著者 | Cheng, Q, Chen, S. | | 登録日 | 2019-04-15 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Genetic and Biochemical Characterization of VMB-1, a Novel Metallo-beta-Lactamase Encoded by a Conjugative, Broad-Host Range IncC Plasmid from Vibrio spp.
Adv Biosyst, 4, 2020
|
|
