5JWO
 
 | | Crystal structure of foldswitch-stabilized KaiB in complex with the N-terminal CI domain of KaiC from Thermosynechococcus elongatus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Circadian clock protein KaiB, Circadian clock protein kinase KaiC | | Authors: | Tseng, R, Goularte, N.F, Chavan, A, Luu, J, Chang, Y, Heilser, J, Tripathi, S, LiWang, A, Partch, C.L. | | Deposit date: | 2016-05-12 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the day-night transition in a bacterial circadian clock.
Science, 355, 2017
|
|
4TKQ
 
 | | Native-SAD phasing for YetJ from Bacillus Subtilis | | Descriptor: | CALCIUM ION, CHLORIDE ION, Uncharacterized protein YetJ | | Authors: | Liu, Q, Chang, Y, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8025 Å) | | Cite: | Multi-crystal native SAD analysis at 6 keV.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7RX9
 
 | | Structure of autoinhibited P-Rex1 | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein, Endolysin chimera, SULFATE ION | | Authors: | Ellisdon, A.M, Chang, Y. | | Deposit date: | 2021-08-22 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structure of the metastatic factor P-Rex1 reveals a two-layered autoinhibitory mechanism.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8T4Y
 
 | | Human HCN1 F186C S264C C309A bound to cAMP, reconstituted in LMNG + SPL | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Burtscher, V, Mount, J, Cowgill, J, Chang, Y, Bickel, K, Yuan, P, Chanda, B. | | Deposit date: | 2023-06-12 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for hyperpolarization-dependent opening of human HCN1 channel.
Nat Commun, 15, 2024
|
|
8T50
 
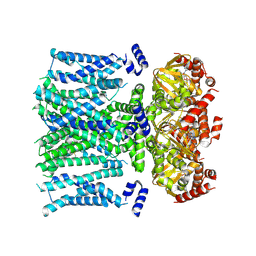 | | Open human HCN1 F186C S264C bound to cAMP, reconstituted in LMNG + SPL | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Burtscher, V, Mount, J, Cowgill, J, Chang, Y, Bickel, K, Yuan, P, Chanda, B. | | Deposit date: | 2023-06-12 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for hyperpolarization-dependent opening of human HCN1 channel.
Nat Commun, 15, 2024
|
|
8T4M
 
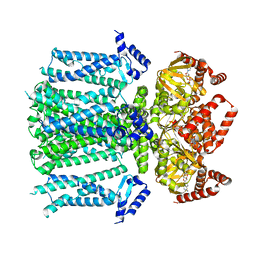 | | Closed human HCN1 F186C S264C bound to cAMP, reconstituted in LMNG + SPL | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Burtscher, V, Mount, J, Cowgill, J, Chang, Y, Bickel, K, Yuan, P, Chanda, B. | | Deposit date: | 2023-06-09 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural basis for hyperpolarization-dependent opening of human HCN1 channel.
Nat Commun, 15, 2024
|
|
5DA4
 
 | |
6NQ8
 
 | |
6NQ9
 
 | |
6NQ7
 
 | |
7UDO
 
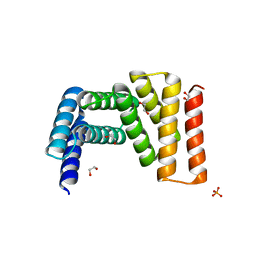 | | Crystal structure of designed helical repeat protein RPB_LRP2_R4 (proteolysis fragment?), forming pseudopolymeric filaments | | Descriptor: | 1,2-ETHANEDIOL, Designed helical repeat protein (DHR) RPB_LRP2_R4, PHOSPHATE ION | | Authors: | Redler, R.L, Chang, Y, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDJ
 
 | | Crystal structure of designed helical repeat protein RPB_PEW3_R4 bound to PAWx4 peptide | | Descriptor: | 4xPAW peptide, De novo designed helical repeat protein RPB_PEW3_R4 | | Authors: | Redler, R.L, Chang, Y, Bhabha, G, Ekiert, D. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
3UCI
 
 | | Crystal structure of Rhodostomin ARLDDL mutant | | Descriptor: | disintegrin | | Authors: | Shiu, J.H, Chen, C.Y, Chen, Y.C, Chang, Y.T, Chang, Y.S, Huang, C.H, Chuang, W.J. | | Deposit date: | 2011-10-27 | | Release date: | 2012-11-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Design of Integrin AlphaVbeta3-Specific Disintegrin for Cancer Therapy
To be Published
|
|
2JW8
 
 | | Solution structure of stereo-array isotope labelled (SAIL) C-terminal dimerization domain of SARS coronavirus nucleocapsid protein | | Descriptor: | Nucleocapsid protein | | Authors: | Takeda, M, Chang, C, Ikeya, T, Guntert, P, Chang, Y, Hsu, Y, Huang, T, Kainosho, M. | | Deposit date: | 2007-10-06 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the c-terminal dimerization domain of SARS coronavirus nucleocapsid protein solved by the SAIL-NMR method
J.Mol.Biol., 380, 2008
|
|
5DA0
 
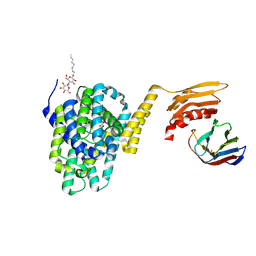 | | Structure of the the SLC26 transporter SLC26Dg in complex with a nanobody | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, Nanobody, Sulphate transporter | | Authors: | Dutzler, R, Geertsma, E.R, Chang, Y, Shaik, F.R. | | Deposit date: | 2015-08-19 | | Release date: | 2015-09-09 | | Last modified: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of a prokaryotic fumarate transporter reveals the architecture of the SLC26 family.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5JWQ
 
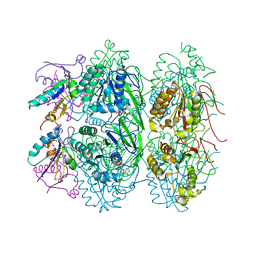 | | Crystal structure of KaiC S431E in complex with foldswitch-stabilized KaiB from Thermosynechococcus elongatus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Circadian clock protein KaiB, Circadian clock protein kinase KaiC | | Authors: | Tseng, R, Goularte, N.F, Chavan, A, Luu, J, Chang, Y, Heilser, J, Tripathi, S, LiWang, A, Partch, C.L. | | Deposit date: | 2016-05-12 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.871 Å) | | Cite: | Structural basis of the day-night transition in a bacterial circadian clock.
Science, 355, 2017
|
|
3OS5
 
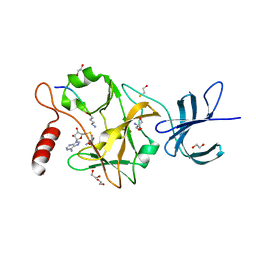 | | SET7/9-Dnmt1 K142me1 complex | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Dnmt1, ... | | Authors: | Esteve, P.-O, Chang, Y, Samaranayake, M, Upadhyay, A.K, Horton, J.R, Feehery, G.R, Cheng, X, Pradhan, S. | | Deposit date: | 2010-09-08 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2MOG
 
 | | Solution structure of the terminal Ig-like domain from Leptospira interrogans LigB | | Descriptor: | Bacterial Ig-like domain, group 2 | | Authors: | Ptak, C.P, Hsieh, C, Lin, Y, Maltsev, A.S, Raman, R, Sharma, Y, Oswald, R.E, Chang, Y. | | Deposit date: | 2014-04-25 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Terminal Immunoglobulin-like Domain from the Leptospira Host-Interacting Outer Membrane Protein, LigB.
Biochemistry, 53, 2014
|
|
2LJV
 
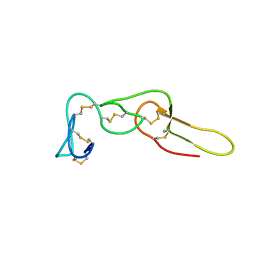 | | Solution structure of Rhodostomin G50L mutant | | Descriptor: | Disintegrin rhodostomin | | Authors: | Chuang, W, Shiu, J, Chen, C, Chen, Y, Chang, Y, Huang, C. | | Deposit date: | 2011-09-29 | | Release date: | 2012-10-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Design of Integrin AlphaVbeta3-Specific Disintegrin for Cancer Therapy
To be Published
|
|
2M75
 
 | |
2M7H
 
 | |
2M7F
 
 | |
7CN9
 
 | | Cryo-EM structure of SARS-CoV-2 Spike ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ho, M, Chang, Y, Wang, C, Wu, Y, Huang, H, Chen, T, Lo, J.M, Chen, X, Ma, C. | | Deposit date: | 2020-07-30 | | Release date: | 2020-08-26 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | A Carbohydrate-Binding Protein from the Edible Lablab Beans Effectively Blocks the Infections of Influenza Viruses and SARS-CoV-2.
Cell Rep, 32, 2020
|
|
4PGV
 
 | |
4PGS
 
 | |
