8WT1
 
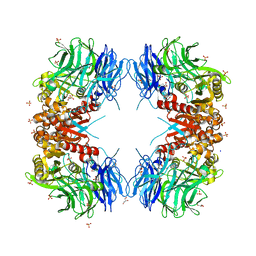 | | Crystal structure of S9 carboxypeptidase from Geobacillus sterothermophilus | | 分子名称: | ALANINE, CITRATE ANION, GLYCEROL, ... | | 著者 | Chandravanshi, K, Kumar, A, Sen, C, Singh, R, Bhange, G.B, Makde, R.D. | | 登録日 | 2023-10-17 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure and solution scattering of Geobacillus stearothermophilus S9 peptidase reveal structural adaptations for carboxypeptidase activity.
Febs Lett., 598, 2024
|
|
7YH4
 
 | |
7Q5N
 
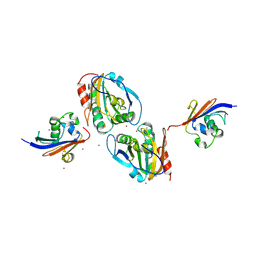 | | Crystal structure of Chaetomium thermophilum Ahp1-Urm1 complex | | 分子名称: | Thioredoxin domain-containing protein, Ubiquitin-related modifier 1, ZINC ION | | 著者 | Ravichandran, K.E, Wilk, P, Grudnik, P, Glatt, S. | | 登録日 | 2021-11-04 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | E2/E3-independent ubiquitin-like protein conjugation by Urm1 is directly coupled to cysteine persulfidation.
Embo J., 41, 2022
|
|
7Q69
 
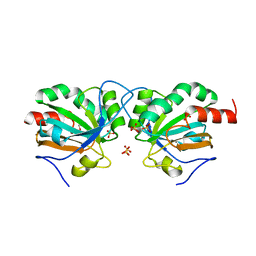 | | Crystal structure of Chaetomium thermophilum C30S Ahp1 in the pre-reaction state | | 分子名称: | GLYCEROL, SULFATE ION, Thioredoxin domain-containing protein | | 著者 | Ravichandran, K.E, Wilk, P, Grudnik, P, Glatt, S. | | 登録日 | 2021-11-05 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | E2/E3-independent ubiquitin-like protein conjugation by Urm1 is directly coupled to cysteine persulfidation.
Embo J., 41, 2022
|
|
7Q6A
 
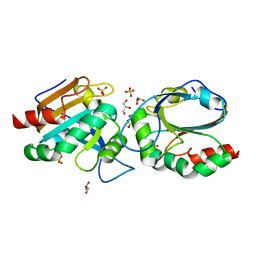 | | Crystal structure of Chaetomium thermophilum C30S Ahp1 in post-reaction state | | 分子名称: | GLYCEROL, SULFATE ION, Thioredoxin domain-containing protein | | 著者 | Ravichandran, K.E, Wilk, P, Grudnik, P, Glatt, S. | | 登録日 | 2021-11-05 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | E2/E3-independent ubiquitin-like protein conjugation by Urm1 is directly coupled to cysteine persulfidation.
Embo J., 41, 2022
|
|
7Q68
 
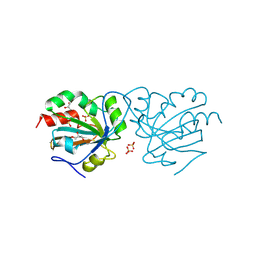 | | Crystal structure of Chaetomium thermophilum wild-type Ahp1 | | 分子名称: | GLYCEROL, SULFATE ION, Thioredoxin domain-containing protein | | 著者 | Ravichandran, K.E, Wilk, P, Grudnik, P, Glatt, S. | | 登録日 | 2021-11-05 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | E2/E3-independent ubiquitin-like protein conjugation by Urm1 is directly coupled to cysteine persulfidation.
Embo J., 41, 2022
|
|
4JDR
 
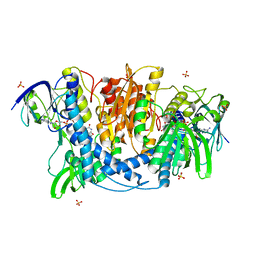 | | Dihydrolipoamide dehydrogenase of pyruvate dehydrogenase from escherichia coli | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Chandrasekhar, K, Arjunan, P, Furey, W. | | 登録日 | 2013-02-25 | | 公開日 | 2013-04-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Insight to the Interaction of the Dihydrolipoamide Acetyltransferase (E2) Core with the Peripheral Components in the Escherichia coli Pyruvate Dehydrogenase Complex via Multifaceted Structural Approaches.
J.Biol.Chem., 288, 2013
|
|
2HPD
 
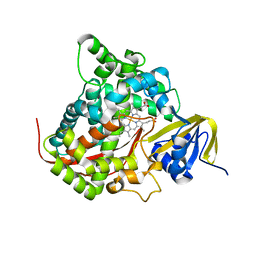 | | CRYSTAL STRUCTURE OF HEMOPROTEIN DOMAIN OF P450BM-3, A PROTOTYPE FOR MICROSOMAL P450'S | | 分子名称: | CYTOCHROME P450 BM-3, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Ravichandran, K.G, Boddupalli, S.S, Hasemann, C.A, Peterson, J.A, Deisenhofer, J. | | 登録日 | 1993-09-16 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of hemoprotein domain of P450BM-3, a prototype for microsomal P450's.
Science, 261, 1993
|
|
4N72
 
 | |
1QC9
 
 | |
2OUL
 
 | | The Structure of Chagasin in Complex with a Cysteine Protease Clarifies the Binding Mode and Evolution of a New Inhibitor Family | | 分子名称: | Chagasin, Falcipain 2 | | 著者 | Wang, S.X, Chand, K, Huang, R, Whisstock, J, Jacobelli, J, Fletterick, R.J, Rosenthal, P.J, McKerrow, J.H. | | 登録日 | 2007-02-11 | | 公開日 | 2008-02-26 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The structure of chagasin in complex with a cysteine protease clarifies the binding mode and evolution of an inhibitor family.
Structure, 15, 2007
|
|
2QTC
 
 | | E. coli Pyruvate dehydrogenase E1 component E401K mutant with phosphonolactylthiamin diphosphate | | 分子名称: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-{(1S)-1-HYDROXY-1-[(R)-HYDROXY(METHOXY)PHOSPHORYL]ETHYL}-5-(2-{[(S)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, MAGNESIUM ION, Pyruvate dehydrogenase E1 component | | 著者 | Furey, W, Arjunan, P, Chandrasekhar, K. | | 登録日 | 2007-08-01 | | 公開日 | 2007-08-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | A Dynamic Loop at the Active Center of the Escherichia coli Pyruvate Dehydrogenase Complex E1 Component Modulates Substrate Utilization and Chemical Communication with the E2 Component
J.Biol.Chem., 282, 2007
|
|
7WF4
 
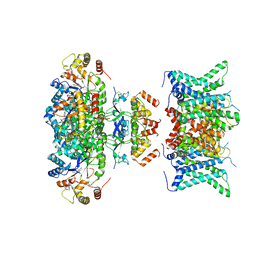 | | Composite map of human Kv1.3 channel in dalazatide-bound state with beta subunits | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3, ... | | 著者 | Tyagi, A, Ahmed, T, Jian, S, Bajaj, S, Ong, S.T, Goay, S.S.M, Zhao, Y, Vorobyov, I, Tian, C, Chandy, K.G, Bhushan, S. | | 登録日 | 2021-12-25 | | 公開日 | 2022-02-09 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Rearrangement of a unique Kv1.3 selectivity filter conformation upon binding of a drug.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WF3
 
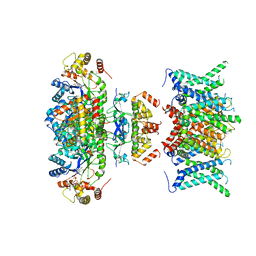 | | Composite map of human Kv1.3 channel in apo state with beta subunits | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3, ... | | 著者 | Tyagi, A, Ahmed, T, Jian, S, Bajaj, S, Ong, S.T, Goay, S.S.M, Zhao, Y, Vorobyov, I, Tian, C, Chandy, K.G, Bhushan, S. | | 登録日 | 2021-12-25 | | 公開日 | 2022-02-09 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Rearrangement of a unique Kv1.3 selectivity filter conformation upon binding of a drug.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5VK2
 
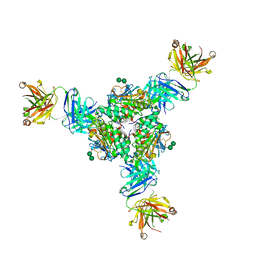 | | Structural basis for antibody-mediated neutralization of Lassa virus | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Hastie, K.M, Zandonatti, M.A, Kleinfelter, L.M, Rowland, M.L, Rowland, M.M, Chandra, K, Branco, L.M, Robinson, J.E, Garry, R.F, Saphire, E.O. | | 登録日 | 2017-04-20 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (3.201 Å) | | 主引用文献 | Structural basis for antibody-mediated neutralization of Lassa virus.
Science, 356, 2017
|
|
4G2K
 
 | | Crystal structure of the Marburg Virus GP2 ectodomain in its post-fusion conformation | | 分子名称: | CHLORIDE ION, GLYCEROL, General control protein GCN4, ... | | 著者 | Malashkevich, V.N, Koellhoffer, J.F, Harrison, J.S, Toro, R, Bhosle, R.C, Chandran, K, Lai, J.R, Almo, S.C. | | 登録日 | 2012-07-12 | | 公開日 | 2012-09-12 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of the Marburg Virus GP2 Core Domain in Its Postfusion Conformation.
Biochemistry, 51, 2012
|
|
1BEI
 
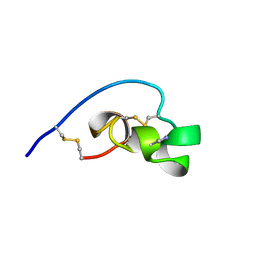 | | Shk-dnp22: A Potent Kv1.3-specific immunosuppressive polypeptide, NMR, 20 structures | | 分子名称: | POTASSIUM CHANNEL TOXIN SHK | | 著者 | Kalman, K, Pennington, M.W, Lanigan, M.D, Nguyen, A, Rauer, H, Mahnir, V, Gutman, G.A, Paschetto, K, Kem, W.R, Grissmer, S, Christian, E.P, Cahalan, M.D, Norton, R.S, Chandy, K.G. | | 登録日 | 1998-05-14 | | 公開日 | 1998-12-02 | | 最終更新日 | 2022-12-21 | | 実験手法 | SOLUTION NMR | | 主引用文献 | ShK-Dap22, a potent Kv1.3-specific immunosuppressive polypeptide.
J.Biol.Chem., 273, 1998
|
|
6A4R
 
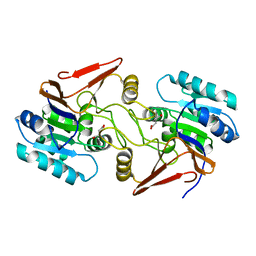 | | Crystal structure of aspartate bound peptidase E from Salmonella enterica | | 分子名称: | ASPARTIC ACID, Peptidase E | | 著者 | Yadav, P, Chandravanshi, K, Goyal, V.D, Singh, R, Kumar, A, Gokhale, S.M, Makde, R.D. | | 登録日 | 2018-06-20 | | 公開日 | 2018-10-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.828 Å) | | 主引用文献 | Structure of Asp-bound peptidase E from Salmonella enterica: Active site at dimer interface illuminates Asp recognition.
FEBS Lett., 592, 2018
|
|
1C2U
 
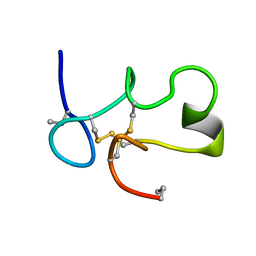 | | SOLUTION STRUCTURE OF [ABU3,35]SHK12-28,17-32 | | 分子名称: | SYNTHETIC PEPTIDE ANALOGUE OF SHK TOXIN | | 著者 | Pennington, M.W, Lanigan, M.D, Kalman, K, Manhir, V.M, Rauer, H, McVaugh, C.T, Behm, D, Donaldson, D, Chandy, K.G, Kem, W.R, Norton, R.S. | | 登録日 | 1999-07-27 | | 公開日 | 1999-11-10 | | 最終更新日 | 2021-11-03 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Role of disulfide bonds in the structure and potassium channel blocking activity of ShK toxin.
Biochemistry, 38, 1999
|
|
6A4S
 
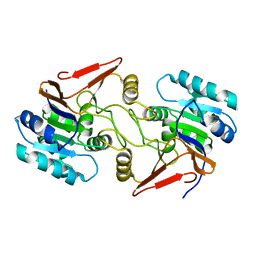 | | Crystal structure of peptidase E with ordered active site loop from Salmonella enterica | | 分子名称: | Peptidase E | | 著者 | Yadav, P, Chandravanshi, K, Goyal, V.D, Singh, R, Kumar, A, Gokhale, S.M, Makde, R.D. | | 登録日 | 2018-06-20 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of Asp-bound peptidase E from Salmonella enterica: Active site at dimer interface illuminates Asp recognition.
FEBS Lett., 592, 2018
|
|
1RN1
 
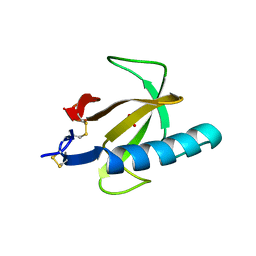 | | THREE-DIMENSIONAL STRUCTURE OF GLN 25-RIBONUCLEASE T1 AT 1.84 ANGSTROMS RESOLUTION: STRUCTURAL VARIATIONS AT THE BASE RECOGNITION AND CATALYTIC SITES | | 分子名称: | RIBONUCLEASE T1 ISOZYME, SULFATE ION | | 著者 | Arni, R.K, Pal, G.P, Ravichandran, K.G, Tulinsky, A, Walz Junior, F.G, Metcalf, P. | | 登録日 | 1991-11-22 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Three-dimensional structure of Gln25-ribonuclease T1 at 1.84-A resolution: structural variations at the base recognition and catalytic sites.
Biochemistry, 31, 1992
|
|
3HTC
 
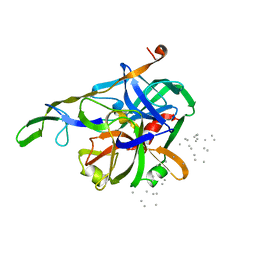 | | THE STRUCTURE OF A COMPLEX OF RECOMBINANT HIRUDIN AND HUMAN ALPHA-THROMBIN | | 分子名称: | ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), HIRUDIN VARIANT 2 | | 著者 | Tulinsky, A, Rydel, T.J, Ravichandran, K.G, Huber, R, Bode, W. | | 登録日 | 1993-06-11 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The structure of a complex of recombinant hirudin and human alpha-thrombin.
Science, 249, 1990
|
|
2G25
 
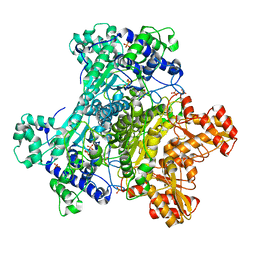 | | E. Coli Pyruvate Dehydrogenase Phosphonolactylthiamin Diphosphate Complex | | 分子名称: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-{(1S)-1-HYDROXY-1-[(R)-HYDROXY(METHOXY)PHOSPHORYL]ETHYL}-5-(2-{[(S)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Furey, W, Arjunan, P, Chandrasekhar, K. | | 登録日 | 2006-02-15 | | 公開日 | 2006-04-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A Thiamin-bound, Pre-decarboxylation Reaction Intermediate Analogue in the Pyruvate Dehydrogenase E1 Subunit Induces Large Scale Disorder-to-Order Transformations in the Enzyme and Reveals Novel Structural Features in the Covalently Bound Adduct.
J.Biol.Chem., 281, 2006
|
|
1OY2
 
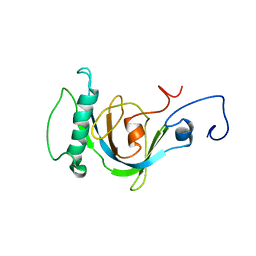 | | Coupling of Folding and Binding in the PTB Domain of the Signaling Protein Shc | | 分子名称: | SHC transforming protein | | 著者 | Farooq, A, Zeng, L, Yan, K.S, Ravichandran, K.S, Zhou, M.-M. | | 登録日 | 2003-04-03 | | 公開日 | 2004-04-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Coupling of Folding and Binding in the PTB Domain of the Signaling Protein Shc
Structure, 11, 2003
|
|
1N3H
 
 | | Coupling of Folding and Binding in the PTB Domain of the Signaling Protein Shc | | 分子名称: | SHC Transforming protein | | 著者 | Farooq, A, Zeng, L, Yan, K.S, Ravichandran, K.S, Zhou, M.-M. | | 登録日 | 2002-10-28 | | 公開日 | 2003-10-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Coupling of Folding and Binding in the PTB Domain of the Signaling Protein Shc
Structure, 11, 2003
|
|
