8QUD
 
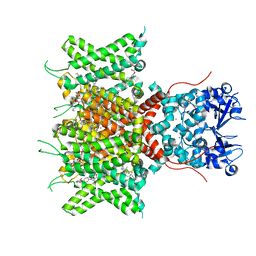 | | Cryo-EM Structure of Human Kv3.1 in Complex with Modulator AUT5 | | Descriptor: | (5R)-5-ethyl-3-(6-spiro[2H-1-benzofuran-3,1'-cyclopropane]-4-yloxypyridin-3-yl)imidazolidine-2,4-dione, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Chi, G, Mckinley, G, Marsden, B, Pike, A.C.W, Ye, M, Brooke, L.M, Bakshi, S, Lakshminarayana, B, Pilati, N, Marasco, A, Gunthorpe, M, Alvaro, G.S, Large, C.H, Williams, E, Sauer, D.B. | | Deposit date: | 2023-10-16 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The binding and mechanism of a positive allosteric modulator of Kv3 channels.
Nat Commun, 15, 2024
|
|
8QUC
 
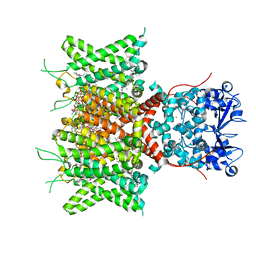 | | Cryo-EM Structure of Human Kv3.1 in Complex with Modulator AUT1 | | Descriptor: | (5R)-5-ethyl-3-[6-(3-methoxy-4-methyl-phenoxy)pyridin-3-yl]imidazolidine-2,4-dione, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Chi, G, Mckinley, G, Marsden, B, Pike, A.C.W, Ye, M, Brooke, L.M, Bakshi, S, Pilati, N, Marasco, A, Gunthorpe, M, Alvaro, G, Large, C, Lakshminaraya, B, Williams, E, Sauer, D.B. | | Deposit date: | 2023-10-16 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The binding and mechanism of a positive allosteric modulator of Kv3 channels.
Nat Commun, 15, 2024
|
|
8QZ2
 
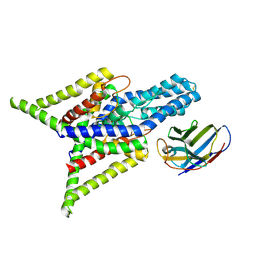 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with an inhibitory nanobody (Nb61) | | Descriptor: | Nanobody 61, POTASSIUM ION, Potassium channel subfamily K member 10 | | Authors: | Baronina, A, Pike, A.C.W, Rodstrom, K.E.J, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
8QZ1
 
 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with a nanobody (Nb58) | | Descriptor: | Isoform B of Potassium channel subfamily K member 10, Nanobody 58, POTASSIUM ION | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Baronina, A, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.588 Å) | | Cite: | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
8QZ3
 
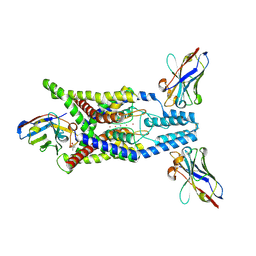 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with an activatory nanobody (Nb67) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Nanobody 67, POTASSIUM ION, ... | | Authors: | Baronina, A, Pike, A.C.W, Rodstrom, K.E.J, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
8QZ4
 
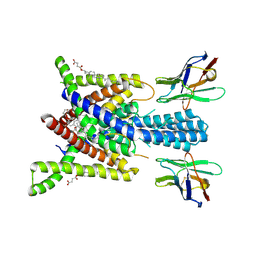 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with an activatory nanobody (Nb76) | | Descriptor: | BARIUM ION, CHOLESTEROL HEMISUCCINATE, Nanobody 76, ... | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Baronina, A, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
4KL1
 
 | | HCN4 CNBD in complex with cGMP | | Descriptor: | ACETATE ION, CYCLIC GUANOSINE MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Lolicato, M, Arrigoni, C, Zucca, S, Nardini, M, Bucchi, A, Schroeder, I, Simmons, K, Bolognesi, M, DiFrancesco, D, Schwede, F, Fishwick, C.W.G, Johnson, A.P.K, Thiel, G, Moroni, A. | | Deposit date: | 2013-05-07 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cyclic dinucleotides bind the C-linker of HCN4 to control channel cAMP responsiveness.
Nat.Chem.Biol., 10, 2014
|
|
5FQ9
 
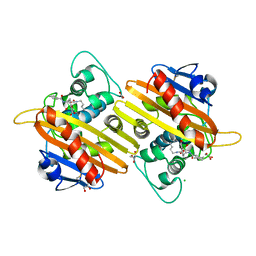 | | Crystal structure of the OXA10 with 1C | | Descriptor: | (3R)-3-(cyclohexylcarbonylamino)-2-oxidanyl-3,4-dihydro-1,2-benzoxaborinine-8-carboxylic acid, ACETATE ION, BETA-LACTAMASE OXA-10, ... | | Authors: | Brem, J, McDonough, M.A, Cain, R, Clifton, I, Fishwick, C.W.G, Schofield, C.J. | | Deposit date: | 2015-12-08 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of metallo-beta-lactamase, serine-beta-lactamase and penicillin-binding protein inhibition by cyclic boronates.
Nat Commun, 7, 2016
|
|
5FV7
 
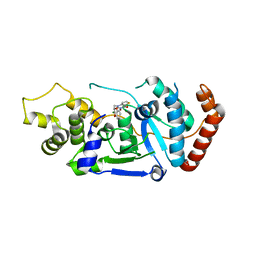 | | Human Fen1 in complex with an N-hydroxyurea compound | | Descriptor: | 1-[(2S)-2,3-dihydro-1,4-benzodioxin-2-ylmethyl]-3-hydroxythieno[3,2-d]pyrimidine-2,4(1H,3H)-dione, FLAP ENDONUCLEASE 1, MAGNESIUM ION | | Authors: | Exell, J.C, Thompson, M.J, Finger, L.D, Shaw, S.K, Abbott, W.M, McWhirter, C, Debreczeni, J.E, Jones, C.D, Nissink, J.W.M, Ward, T.A, Sioberg, C.W.L, Molina, D.M, Durant, S.T, Grasby, J.A. | | Deposit date: | 2016-02-03 | | Release date: | 2016-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Cellular Active N-Hydroxyurea Fen1 Inhibitors Block Substrate Entry to the Active Site
Nat.Chem.Biol., 12, 2016
|
|
3DA2
 
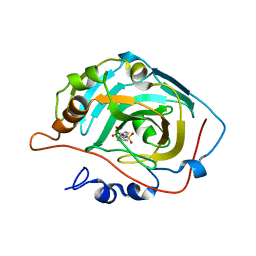 | | X-ray structure of human carbonic anhydrase 13 in complex with inhibitor | | Descriptor: | CHLORIDE ION, Carbonic anhydrase 13, N-(4-chlorobenzyl)-N-methylbenzene-1,4-disulfonamide, ... | | Authors: | Pilka, E.S, Picaud, S.S, Yue, W.W, King, O.N.F, Bray, J.E, Filippakopoulos, P, Roos, A.K, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Wikstrom, M, Edwards, A.M, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-28 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray structure of human carbonic anhydrase 13 in complex with inhibitor.
To be Published
|
|
3QK3
 
 | | Crystal structure of human beta-crystallin B3 | | Descriptor: | 1,2-ETHANEDIOL, Beta-crystallin B3, SULFATE ION | | Authors: | Krojer, T, Gileadi, C, Cocking, R, Muniz, J, Pilka, E, Yue, W.W, Vollmar, M, Pike, A.C.W, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: |
|
|
6RV2
 
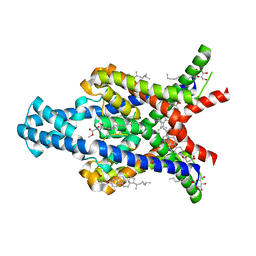 | | Crystal structure of the human two pore domain potassium ion channel TASK-1 (K2P3.1) in a closed conformation | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Zhang, W, Quigley, A, Speedman, D, Mukhopadhyay, S.M.M, Shrestha, L, Chalk, R, Venkaya, S, Bushell, S.R, Tessitore, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-30 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A lower X-gate in TASK channels traps inhibitors within the vestibule.
Nature, 582, 2020
|
|
6S3Q
 
 | | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with TFB-TBOA | | Descriptor: | (2~{S},3~{S})-2-azanyl-3-[[3-[[4-(trifluoromethyl)phenyl]carbonylamino]phenyl]methoxy]butanedioic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Baronina, A, Pike, A.C.W, Yu, X, Dong, Y.Y, Shintre, C.A, Tessitore, A, Chu, A, Rotty, B, Venkaya, S, Mukhopadhyay, S, Borkowska, O, Chalk, R, Shrestha, L, Burgess-Brown, N.A, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Han, S, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structure of human excitatory amino acid transporter 3 (EAAT3)
TO BE PUBLISHED
|
|
6S4L
 
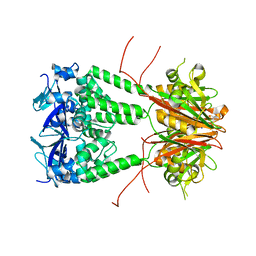 | | Structure of human KCTD1 | | Descriptor: | BTB/POZ domain-containing protein KCTD1, IODIDE ION, SODIUM ION | | Authors: | Pinkas, D.M, Bufton, J.C, Fox, A.E, Pike, A.C.W, Newman, J.A, Krojer, T, Shrestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure of human KCTD1
To be published
|
|
3EMW
 
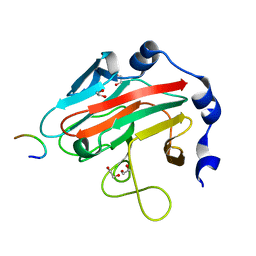 | | Crystal Structure of human splA/ryanodine receptor domain and SOCS box containing 2 (SPSB2) in complex with a 20-residue VASA peptide | | Descriptor: | 1,2-ETHANEDIOL, Peptide (VASA), SPRY domain-containing SOCS box protein 2 | | Authors: | Filippakopoulos, P, Sharpe, T, Keates, T, Murray, J.W, Savitsky, P, Roos, A.K, Pike, A.C.W, Von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for Par-4 recognition by the SPRY domain- and SOCS box-containing proteins SPSB1, SPSB2, and SPSB4.
J.Mol.Biol., 401, 2010
|
|
6S4M
 
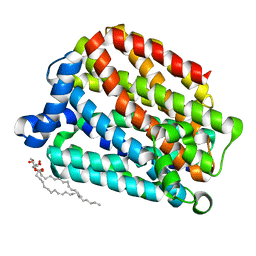 | | Crystal structure of the human organic anion transporter MFSD10 (TETRAN) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRIC ACID, Major facilitator superfamily domain-containing protein 10 | | Authors: | Pascoa, T.C, Pike, A.C.W, Bushell, S.R, Quigley, A, Chu, A, Mukhopadhyay, S.M.M, Shrestha, L, Venkaya, S, Chalk, R, Burgess-Brown, N.A, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-28 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human organic anion transporter TETRAN (MFSD10)
To be published
|
|
3F2N
 
 | | Crystal Structure of Human Haspin with an Imidazo-pyridazine ligand | | Descriptor: | (2S)-2-{[3-(3-aminophenyl)imidazo[1,2-b]pyridazin-6-yl]amino}-3-methylbutan-1-ol, 1,2-ETHANEDIOL, PHOSPHATE ION, ... | | Authors: | Filippakopoulos, P, Eswaran, J, Keates, T, Burgess-Brown, N, Fedorov, O, Yue, W.W, Murray, J.W, Pike, A.C.W, Von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Human Haspin with an Imidazo-pyridazine ligand
To be Published
|
|
3F3S
 
 | | The Crystal Structure of Human Lambda-Crystallin, CRYL1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Lambda-crystallin homolog, ... | | Authors: | Ugochukwu, E, Johansson, C, Yue, W.W, Kochan, G, Pilka, E, Kramm, A, Pike, A.C.W, Filippakopoulos, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-31 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human Lambda-Crystallin, CRYL1
To be Published
|
|
3CP6
 
 | | Crystal structure of human farnesyl diphosphate synthase (T201A mutant) complexed with Mg and biphosphonate inhibitor | | Descriptor: | (4aS,7aR)-octahydro-1H-cyclopenta[b]pyridine-6,6-diylbis(phosphonic acid), Farnesyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Pilka, E.S, Dunford, J.E, Guo, K, Pike, A.C.W, von Delft, F, Barnett, B.L, Ebetino, F.H, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Russell, R.G.G, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-31 | | Release date: | 2008-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Human farnesyl diphosphate synthase (T201A mutant) complexed with Mg and biphosphonate inhibitor.
To be Published
|
|
3CY3
 
 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and the JNK inhibitor V | | Descriptor: | (2S)-1,3-benzothiazol-2-yl{2-[(2-pyridin-3-ylethyl)amino]pyrimidin-4-yl}ethanenitrile, 1,2-ETHANEDIOL, Pimtide peptide, ... | | Authors: | Filippakopoulos, P, Bullock, A, Fedorov, O, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and the JNK inhibitor V.
To be Published
|
|
1BOY
 
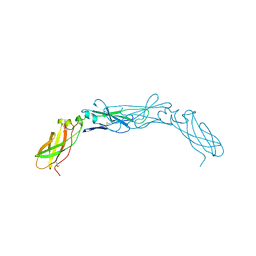 | |
3CY2
 
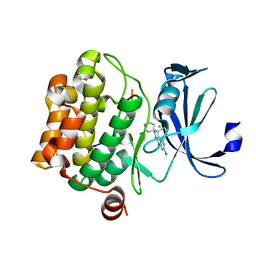 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a beta carboline ligand II | | Descriptor: | (4R)-7-chloro-9-methyl-1-oxo-1,2,4,9-tetrahydrospiro[beta-carboline-3,4'-piperidine]-4-carbonitrile, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Filippakopoulos, P, Bullock, A, Fedorov, O, Huber, K, Bracher, F, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | 7,8-Dichloro-1-oxo-beta-carbolines as a Versatile Scaffold for the Development of Potent and Selective Kinase Inhibitors with Unusual Binding Modes
J.Med.Chem., 55, 2012
|
|
3ZDQ
 
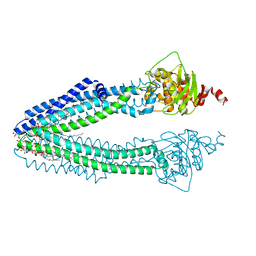 | | STRUCTURE OF THE HUMAN MITOCHONDRIAL ABC TRANSPORTER, ABCB10 (NUCLEOTIDE-FREE FORM) | | Descriptor: | ATP-BINDING CASSETTE SUB-FAMILY B MEMBER 10, MITOCHONDRIAL, CARDIOLIPIN, ... | | Authors: | Pike, A.C.W, Shintre, C.A, Krojer, T, von Delft, F, Vollmar, M, Mukhopadhyay, S, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Carpenter, E.P. | | Deposit date: | 2012-11-30 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of Abcb10, a Human ATP-Binding Cassette Transporter in Apo- and Nucleotide-Bound States
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6R65
 
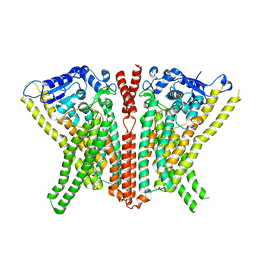 | | Crystal Structure of human TMEM16K / Anoctamin 10 (Form 2) | | Descriptor: | Anoctamin-10, CALCIUM ION | | Authors: | Bushell, S.R, Pike, A.C.W, Chu, A, Tessitore, A, Rotty, B, Mukhopadhyay, S, Kupinska, K, Shrestha, L, Borkowska, O, Chalk, R, Burgess-Brown, N.A, Love, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-26 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
6R7Z
 
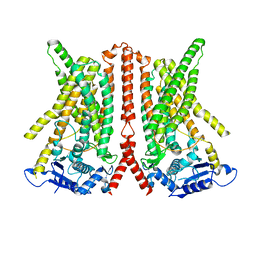 | | CryoEM structure of calcium-free human TMEM16K / Anoctamin 10 in detergent (closed form) | | Descriptor: | Anoctamin-10 | | Authors: | Pike, A.C.W, Bushell, S.R, Shintre, C.A, Tessitore, A, Chu, A, Mukhopadhyay, S, Shrestha, L, Chalk, R, Burgess-Brown, N.A, Love, J, Huiskonen, J.T, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-29 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.14 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
