7T91
 
 | |
1S1T
 
 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with UC-781 | | Descriptor: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, PHOSPHATE ION, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
1RVS
 
 | | STRUCTURE OF TRANSTHYRETIN IN AMYLOID FIBRILS DETERMINED BY SOLID-STATE MAGIC ANGLE SPINNING NMR | | Descriptor: | Transthyretin | | Authors: | Jaroniec, C.P, Macphee, C.E, Bajaj, V.S, Mcmahon, M.T, Dobson, C.M, Griffin, R.G. | | Deposit date: | 2003-12-14 | | Release date: | 2004-01-20 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution molecular structure of a peptide in an amyloid fibril determined by magic angle spinning NMR spectroscopy
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
7UYU
 
 | | Crystal structure of TYK2 kinase domain in complex with compound 30 | | Descriptor: | 2-(2,6-difluorophenyl)-4-[4-(pyrrolidine-1-carbonyl)anilino]-5H-pyrrolo[3,4-b]pyridin-5-one, Non-receptor tyrosine-protein kinase TYK2, SULFATE ION | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYR
 
 | | Crystal structure of TYK2 kinase domain in complex with compound 12 | | Descriptor: | 2-(4-{[2-(2,6-difluorophenyl)-5-oxo-5H-pyrrolo[3,4-d]pyrimidin-4-yl]amino}phenyl)-N-ethylacetamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYW
 
 | | Crystal structure of JAK2 kinase domain in complex with compound 30 | | Descriptor: | 2-(2,6-difluorophenyl)-4-[4-(pyrrolidine-1-carbonyl)anilino]-5H-pyrrolo[3,4-b]pyridin-5-one, Tyrosine-protein kinase JAK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYT
 
 | | Crystal structure of TYK2 kinase domain in complex with compound 25 | | Descriptor: | 6-{[(2M)-2-(2-chloro-6-fluorophenyl)-5-oxo-5H-pyrrolo[3,4-b]pyridin-4-yl]amino}-N-ethylpyridine-3-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYV
 
 | | Crystal structure of JAK3 kinase domain in complex with compound 25 | | Descriptor: | 6-{[(2M)-2-(2-chloro-6-fluorophenyl)-5-oxo-5H-pyrrolo[3,4-b]pyridin-4-yl]amino}-N-ethylpyridine-3-carboxamide, CHLORIDE ION, Tyrosine-protein kinase JAK3 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7UYS
 
 | | Crystal structure of TYK2 kinase domain in complex with compound 16 | | Descriptor: | 2-(2,6-difluorophenyl)-4-(4-methoxyanilino)-5H-pyrrolo[3,4-d]pyrimidin-5-one, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
6U1T
 
 | | Crystal structure of anti-Nipah virus (NiV) F 5B3 antibody Fab fragment | | Descriptor: | CHLORIDE ION, antigen-binding (Fab) fragment, heavy chain, ... | | Authors: | Dang, H.V, Chan, Y.P, Park, Y.J, Snijder, J, Da Silva, S.C, Vu, B, Yan, L, Feng, Y.R, Rockx, B, Geisbert, T, Mire, C, Mire, C.E, BBroder, C.C, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-08-16 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.483 Å) | | Cite: | An antibody against the F glycoprotein inhibits Nipah and Hendra virus infections.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6TYS
 
 | | A potent cross-neutralizing antibody targeting the fusion glycoprotein inhibits Nipah virus and Hendra virus infection | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5B3 antibody heavy chain, ... | | Authors: | Dang, H.V, Chan, Y.P, Park, Y.J, Snijder, J, Da Silva, S.C, Vu, B, Yan, L, Feng, Y.R, Rockx, B, Geisbert, T, Mire, C.E, Broder, C.B, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-08-09 | | Release date: | 2019-10-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An antibody against the F glycoprotein inhibits Nipah and Hendra virus infections.
Nat.Struct.Mol.Biol., 26, 2019
|
|
8F2P
 
 | |
1EHN
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT E315Q COMPLEXED WITH OCTA-N-ACETYLCHITOOCTAOSE (NAG)8. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-02-22 | | Release date: | 2001-02-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
2RCN
 
 | |
4ZYY
 
 | | Human GAR transformylase in complex with GAR and (S)-2-({5-[4-(2-Amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]-pyrimidin-6-yl)butyl]thiophene-3-carbonyl}amino)pentanedioic acid (AGF118) | | Descriptor: | (S)-2-({5-[4-(2-Amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]-pyrimidin-6-yl)butyl]thiophene-3-carbonyl}amino)pentanedioic acid, GLYCINAMIDE RIBONUCLEOTIDE, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Deis, S.M, Dann III, C.E. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Enzymatic Analysis of Tumor-Targeted Antifolates That Inhibit Glycinamide Ribonucleotide Formyltransferase.
Biochemistry, 55, 2016
|
|
4ZZ3
 
 | | Human GAR transformylase in complex with GAR and pemetrexed | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{4-[2-(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)ethyl]benzoyl}-L-glutamic acid, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Deis, S.M, Dann III, C.E. | | Deposit date: | 2015-05-22 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | 6-Substituted Pyrrolo[2,3-d]pyrimidine Thienoyl Regioisomers as Targeted Antifolates for Folate Receptor alpha and the Proton-Coupled Folate Transporter in Human Tumors.
J.Med.Chem., 58, 2015
|
|
8FFJ
 
 | | Structure of Zanidatamab bound to HER2 | | Descriptor: | Receptor tyrosine-protein kinase erbB-2, Zanidatamab Heavy Chain A, Zanidatamab Heavy Chain B, ... | | Authors: | Worrall, L.J, Atkinson, C.E, Sanches, M, Dixit, S, Strynadka, N.C.J. | | Deposit date: | 2022-12-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | An anti-HER2 biparatopic antibody that induces unique HER2 clustering and complement-dependent cytotoxicity.
Nat Commun, 14, 2023
|
|
4PXZ
 
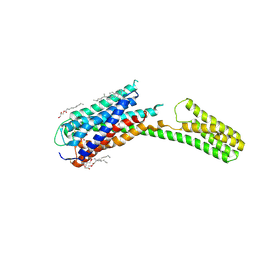 | | Crystal structure of P2Y12 receptor in complex with 2MeSADP | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(methylsulfanyl)adenosine 5'-(trihydrogen diphosphate), CHOLESTEROL, ... | | Authors: | Zhang, J, Zhang, K, Gao, Z.G, Paoletta, S, Zhang, D, Han, G.W, Li, T, Ma, L, Zhang, W, Muller, C.E, Yang, H, Jiang, H, Cherezov, V, Katritch, V, Jacobson, K.A, Stevens, R.C, Wu, B, Zhao, Q, GPCR Network (GPCR) | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Agonist-bound structure of the human P2Y12 receptor
Nature, 509, 2014
|
|
8FON
 
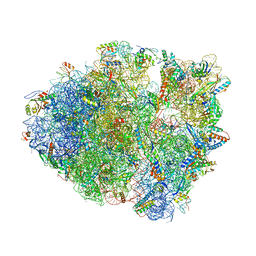 | | Crystal structure of tRNA^Lys(SUU) bound to AUA codon in the ribosomal P site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Nguyen, H.A, Hoffer, E.D, Maehigashi, T, Fagan, C.E, Dunham, C.M. | | Deposit date: | 2023-01-02 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Structural basis for reduced ribosomal A-site fidelity in response to P-site codon-anticodon mismatches.
J.Biol.Chem., 299, 2023
|
|
8FOM
 
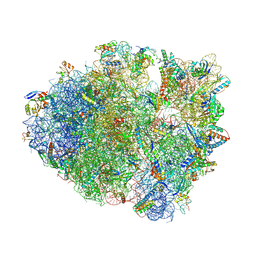 | | Crystal structure of tRNA^Lys(SUU) bound to UAA codon in the ribosomal P site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Nguyen, H.A, Hoffer, E.D, Maehigashi, T, Fagan, C.E, Dunham, C.M. | | Deposit date: | 2023-01-02 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-26 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Structural basis for reduced ribosomal A-site fidelity in response to P-site codon-anticodon mismatches.
J.Biol.Chem., 299, 2023
|
|
8FWN
 
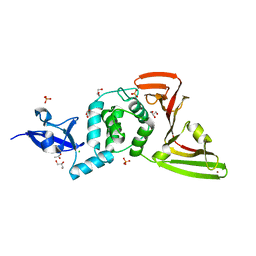 | | Crystal structure of SARS-CoV-2 papain-like protease C111S mutant | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Bezerra, E.H.S, Soprano, A.S, Tonoli, C.C.C, Prado, P.F.V, da Silva, J.C, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | Deposit date: | 2023-01-23 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of SARS-CoV-2 papain-like protease C111S mutant
To Be Published
|
|
1FFR
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT Y390F COMPLEXED WITH HEXA-N-ACETYLCHITOHEXAOSE (NAG)6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-07-26 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
2UXE
 
 | | The structure of Vaccinia virus N1 | | Descriptor: | HYPOTHETICAL PROTEIN | | Authors: | Cooray, S, Bahar, M.W, Abrescia, N.G.A, McVey, C.E, Bartlett, N.W, Chen, R.A.-J, Stuart, D.I, Grimes, J.M, Smith, G.L. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Functional and Structural Studies of the Vaccinia Virus Virulence Factor N1 Reveal a Bcl-2-Like Anti- Apoptotic Protein
J.Gen.Virol., 88, 2007
|
|
4M3P
 
 | | Betaine-Homocysteine S-Methyltransferase from Homo sapiens complexed with Homocysteine | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Betaine--homocysteine S-methyltransferase 1, POTASSIUM ION, ... | | Authors: | Koutmos, M, Yamada, K, Mladkova, J, Paterova, J, Diamond, C.E, Tryon, K, Jungwirth, P, Garrow, T.A, Jiracek, J. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Specific potassium ion interactions facilitate homocysteine binding to betaine-homocysteine S-methyltransferase.
Proteins, 82, 2014
|
|
8FID
 
 | | Crystal Structure of Erwinia tracheiphila CYP114 in complex with ent-kaurenoic acid (Crystal Form 2) | | Descriptor: | (8alpha,9beta,10alpha,13alpha)-kaur-16-en-18-oic acid, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Stewart Jr, C.E, Nagel, R. | | Deposit date: | 2022-12-16 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Dual factors required for cytochrome-P450-mediated hydrocarbon ring contraction in bacterial gibberellin phytohormone biosynthesis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
