2YGT
 
 | | Clostridium perfringens delta-toxin | | Descriptor: | DELTA TOXIN, GLYCEROL, IMIDAZOLE, ... | | Authors: | Huyet, J, Naylor, C.E, Gibert, M, Popoff, M.R, Basak, A.K. | | Deposit date: | 2011-04-20 | | Release date: | 2012-05-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights Into Clostridium Perfringens Delta Toxin Pore Formation.
Plos One, 8, 2013
|
|
3B66
 
 | | Crystal structure of the androgen receptor ligand binding domain in complex with SARM S-21 | | Descriptor: | 4-{[(1R,2S)-1,2-dihydroxy-2-methyl-3-(4-nitrophenoxy)propyl]amino}-2-(trifluoromethyl)benzonitrile, Androgen receptor | | Authors: | Bohl, C.E, Miller, D.D, Dalton, J.T. | | Deposit date: | 2007-10-27 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Effect of B-ring substitution pattern on binding mode of propionamide selective androgen receptor modulators
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3B9J
 
 | | Structure of Xanthine Oxidase with 2-hydroxy-6-methylpurine | | Descriptor: | 6-methyl-3,9-dihydro-2H-purin-2-one, CALCIUM ION, DIOXOTHIOMOLYBDENUM(VI) ION, ... | | Authors: | Pauff, J.M, Zhang, J, Bell, C.E, Hille, R. | | Deposit date: | 2007-11-05 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate orientation in xanthine oxidase: crystal structure of enzyme in reaction with 2-hydroxy-6-methylpurine.
J.Biol.Chem., 283, 2008
|
|
8RTZ
 
 | | The structure of E. coli penicillin binding protein 3 (PBP3) in complex with a bicyclic peptide inhibitor | | Descriptor: | 1,1',1''-(1,3,5-triazinane-1,3,5-triyl)tripropan-1-one, Bicyclic peptide inhibitor, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Newman, H, Rowland, C.E, Dods, R, Lewis, N, Stanway, S.J, Bellini, D, Beswick, P. | | Deposit date: | 2024-01-29 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery and chemical optimisation of a Potent, Bi-cyclic (Bicycle) Antimicrobial Inhibitor of Escherichia coli PBP3
To Be Published
|
|
6B72
 
 | | A novel HIV-1 Nef dimer interface induced by a single octyl-glucoside molecule | | Descriptor: | Protein Nef, octyl beta-D-glucopyranoside | | Authors: | Wu, M, Augelli-Szafran, C.E, Ptak, R.G, Smithgall, T.E. | | Deposit date: | 2017-10-03 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A single beta-octyl glucoside molecule induces HIV-1 Nef dimer formation in the absence of partner protein binding.
PLoS ONE, 13, 2018
|
|
6BNZ
 
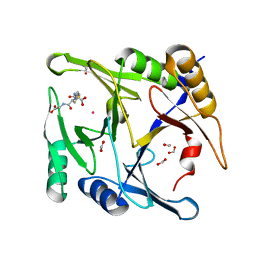 | | Crystal structure of E144Q-glyoxalase I mutant from Zea mays in space group P4(1)2(1)2 | | Descriptor: | COBALT (II) ION, FORMIC ACID, GLUTATHIONE, ... | | Authors: | Alvarez, C.E, Agostini, R.B, Gonzalez, J.M, Drincovich, M.F, Campos Bermudez, V.A, Klinke, S. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Deciphering the number and location of active sites in the monomeric glyoxalase I of Zea mays.
Febs J., 286, 2019
|
|
6C42
 
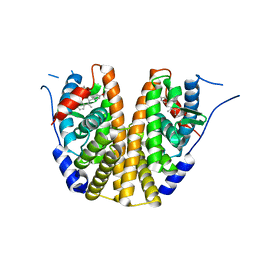 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with OP1156 | | Descriptor: | (2R,3S,4R)-3-(4-hydroxyphenyl)-4-methyl-2-{4-[2-(pyrrolidin-1-yl)ethoxy]phenyl}-3,4-dihydro-2H-1-benzopyran-7-ol, Estrogen receptor | | Authors: | Fanning, S.W, Hodges-Gallager, L, Myles, D.C, Sun, R, Fowler, C.E, Green, B.D, Harmon, C.L, Greene, G.L, Kushner, P.J. | | Deposit date: | 2018-01-11 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Specific stereochemistry of OP-1074 disrupts estrogen receptor alpha helix 12 and confers pure antiestrogenic activity.
Nat Commun, 9, 2018
|
|
3B67
 
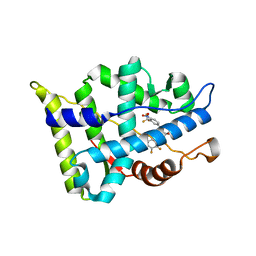 | |
3B65
 
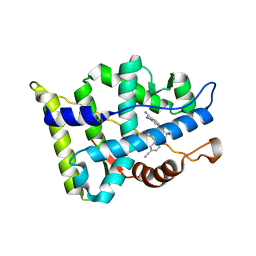 | |
8RJZ
 
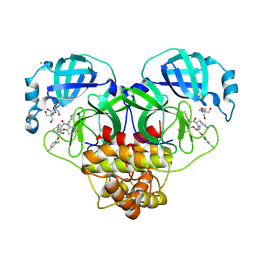 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GUE-3801 (compound 80 in publication) | | Descriptor: | (7~{S})-6-[2-[2,4-bis(chloranyl)phenoxy]ethanoyl]-14-fluoranyl-10-(iminomethyl)-9-methyl-7-(phenylmethyl)-2-oxa-6,9,10-triazabicyclo[10.4.0]hexadeca-1(12),13,15-trien-8-one, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase nsp5, ... | | Authors: | Strater, N, Claff, T, Sylvester, K, Mueller, C.E, Guetschow, M, Useini, A. | | Deposit date: | 2023-12-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Macrocyclic Azapeptide Nitriles: Structure-Based Discovery of Potent SARS-CoV-2 Main Protease Inhibitors as Antiviral Drugs.
J.Med.Chem., 67, 2024
|
|
8RJY
 
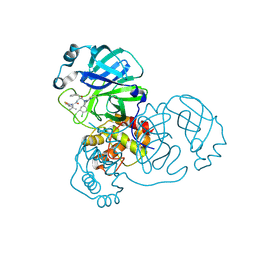 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalent inhibitor GUE-3899 (compound 58 in publication) | | Descriptor: | 3C-like proteinase nsp5, ~{N}-[(2~{S})-1-[[(2~{S})-1-[[(4-chlorophenyl)methyl-(iminomethyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]amino]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]thiophene-2-carboxamide | | Authors: | Strater, N, Claff, T, Sylvester, K, Mueller, C.E, Guetschow, M, Useini, A. | | Deposit date: | 2023-12-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Macrocyclic Azapeptide Nitriles: Structure-Based Discovery of Potent SARS-CoV-2 Main Protease Inhibitors as Antiviral Drugs.
J.Med.Chem., 67, 2024
|
|
3C9D
 
 | | Crystal structure of Vps75 | | Descriptor: | Vacuolar protein sorting-associated protein 75 | | Authors: | Berndsen, C.E, Tsubota, T, Lindner, S.E, Lee, S, Holton, J.M, Kaufman, P.D, Keck, J.L, Denu, J.M. | | Deposit date: | 2008-02-15 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular functions of the histone acetyltransferase chaperone complex Rtt109-Vps75
Nat.Struct.Mol.Biol., 15, 2008
|
|
8RJV
 
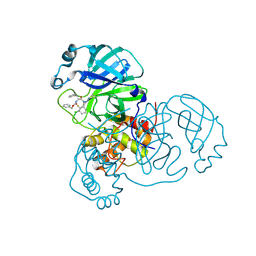 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalent inhibitor GUE-3778 (compound 12 in publication) | | Descriptor: | (phenylmethyl) ~{N}-[(2~{S})-1-[[(3-chloranyl-2-fluoranyl-phenyl)methyl-(iminomethyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]carbamate, 3C-like proteinase nsp5 | | Authors: | Strater, N, Claff, T, Sylvester, K, Mueller, C.E, Guetschow, M, Useini, A. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Macrocyclic Azapeptide Nitriles: Structure-Based Discovery of Potent SARS-CoV-2 Main Protease Inhibitors as Antiviral Drugs.
J.Med.Chem., 67, 2024
|
|
6BZ8
 
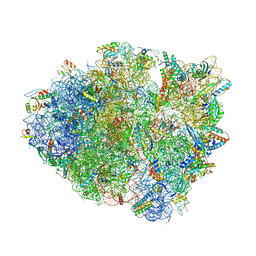 | | Thermus thermophilus 70S containing 16S G347U point mutation and near-cognate ASL Leucine in A site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hoffer, E.D, Maehigashi, T, Fagan, C.E, Dunham, C.M. | | Deposit date: | 2017-12-22 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.74 Å) | | Cite: | Ribosomal ambiguity (ram) mutations promote the open (off) to closed (on) transition and thereby increase miscoding.
Nucleic Acids Res., 47, 2019
|
|
8RLN
 
 | | Crystal structure of human adenosine A2A receptor (construct A2A-PSB2-bRIL) complexed with the partial antagonist LUF5834 at the orthosteric pocket | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-azanyl-4-(4-hydroxyphenyl)-6-(1~{H}-imidazol-2-ylmethylsulfanyl)pyridine-3,5-dicarbonitrile, ... | | Authors: | Strater, N, Claff, T, Weisse, R.H, Muller, C.E. | | Deposit date: | 2024-01-03 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural Insights into Partial Activation of the Prototypic G Protein-Coupled Adenosine A 2A Receptor.
Acs Pharmacol Transl Sci, 7, 2024
|
|
4LMF
 
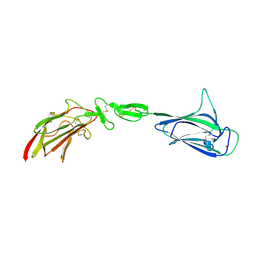 | | C1s CUB1-EGF-CUB2 | | Descriptor: | CALCIUM ION, Complement C1s subcomponent heavy chain, SODIUM ION | | Authors: | Wallis, R, Venkatraman Girija, U, Moody, P.C.E, Marshall, J.E. | | Deposit date: | 2013-07-10 | | Release date: | 2013-08-07 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.921 Å) | | Cite: | Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6BUW
 
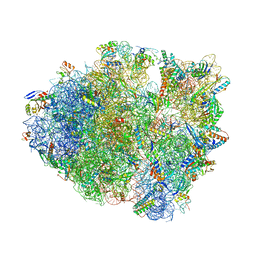 | | Thermus thermophilus 70S complex containing 16S G299A ram mutation and empty A site. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hoffer, E.D, Maehigashi, T, Fagan, C.E, Dunham, C.M. | | Deposit date: | 2017-12-11 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Ribosomal ambiguity (ram) mutations promote the open (off) to closed (on) transition and thereby increase miscoding.
Nucleic Acids Res., 47, 2019
|
|
4M1P
 
 | |
8QDC
 
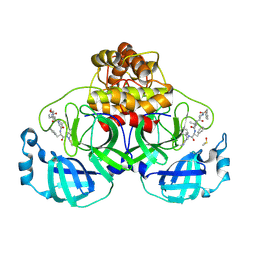 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalent inhibitor GUE-3642 (compound 1 in publication) | | Descriptor: | (phenylmethyl) ~{N}-[(2~{S})-1-[[(2~{S})-1-[[(2~{S})-1-[[iminomethyl-(phenylmethyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]amino]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]amino]-1-oxidanylidene-butan-2-yl]carbamate, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Strater, N, Claff, T, Sylvester, K, Mueller, C.E, Guetschow, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Development of an active-site titrant for SARS-CoV-2 main protease as an indispensable tool for evaluating enzyme kinetics.
Acta Pharm Sin B, 14, 2024
|
|
6CUZ
 
 | | Engineered TrpB from Pyrococcus furiosus, PfTrpB7E6 with (2S,3R)-ethylserine bound as the amino-acrylate | | Descriptor: | (2E)-2-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]pent-2-enoic acid, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Scheele, R.A, Buller, A.R, Boville, C.E, Arnold, F.H. | | Deposit date: | 2018-03-27 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineered Biosynthesis of beta-Alkyl Tryptophan Analogues.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
3BEO
 
 | | A Structural Basis for the allosteric regulation of non-hydrolyzing UDP-GlcNAc 2-epimerases | | Descriptor: | UDP-N-acetylglucosamine 2-epimerase, URIDINE-5'-DIPHOSPHATE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Velloso, L.M, Bhaskaran, S.S, Schuch, R, Fischetti, V.A, Stebbins, C.E. | | Deposit date: | 2007-11-19 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structural basis for the allosteric regulation of non-hydrolysing UDP-GlcNAc 2-epimerases.
Embo Rep., 9, 2008
|
|
3B5R
 
 | |
6BNX
 
 | | Crystal structure of V278E-glyoxalase I mutant from Zea mays in space group P6(3) | | Descriptor: | COBALT (II) ION, Lactoylglutathione lyase | | Authors: | Alvarez, C.E, Agostini, R.B, Gonzalez, J.M, Drincovich, M.F, Campos Bermudez, V.A, Klinke, S. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Deciphering the number and location of active sites in the monomeric glyoxalase I of Zea mays.
Febs J., 286, 2019
|
|
4LOS
 
 | | C1s CUB2-CCP1 | | Descriptor: | CALCIUM ION, Complement C1s subcomponent heavy chain | | Authors: | Wallis, R, Venkatraman Girija, U, Moody, P.C.E, Marshall, J.E, Gingras, A.R. | | Deposit date: | 2013-07-13 | | Release date: | 2013-08-07 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LOT
 
 | | C1s CUB2-CCP1-CCP2 | | Descriptor: | Complement C1s subcomponent heavy chain | | Authors: | Wallis, R, Venkatraman Girija, U, Moody, P.C.E, Marshall, J.E. | | Deposit date: | 2013-07-13 | | Release date: | 2013-08-07 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
