1J3G
 
 | | Solution structure of Citrobacter Freundii AmpD | | Descriptor: | AmpD protein, ZINC ION | | Authors: | Liepinsh, E, Genereux, C, Dehareng, D, Joris, B, Otting, G. | | Deposit date: | 2003-01-31 | | Release date: | 2003-02-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Citrobacter freundii AmpD, Comparison with Bacteriophage T7 Lysozyme
and Homology with PGRP Domains
J.Mol.Biol., 327, 2003
|
|
3D89
 
 | | Crystal Structure of a Soluble Rieske Ferredoxin from Mus musculus | | Descriptor: | 1,2-ETHANEDIOL, FE2/S2 (INORGANIC) CLUSTER, Rieske domain-containing protein | | Authors: | Levin, E.J, McCoy, J.G, Elsen, N.L, Seder, K.D, Bingman, C.A, Wesenberg, G.E, Fox, B.G, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2008-05-22 | | Release date: | 2008-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.071 Å) | | Cite: | X-ray structure of a soluble Rieske-type ferredoxin from Mus musculus.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3M7M
 
 | | Crystal structure of monomeric hsp33 | | Descriptor: | 33 kDa chaperonin | | Authors: | Chi, S.W, Jeong, D.G, Woo, J.R, Park, B.C, Ryu, S.E, Kim, S.J. | | Deposit date: | 2010-03-16 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of monomeric hsp33
To be Published
|
|
1J4O
 
 | | REFINED NMR STRUCTURE OF THE FHA1 DOMAIN OF YEAST RAD53 | | Descriptor: | PROTEIN KINASE SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-03 | | Release date: | 2001-12-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
3M6D
 
 | | The crystal structure of the d307a mutant of glycoside Hydrolase (family 31) from ruminococcus obeum atcc 29174 | | Descriptor: | Uncharacterized protein | | Authors: | Tan, K, Tesar, C, Freeman, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-15 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Novel alpha-glucosidase from human gut microbiome: substrate specificities and their switch.
Faseb J., 24, 2010
|
|
8RT0
 
 | | BTV-15 VP5 pH 6.0 | | Descriptor: | 1,2-ETHANEDIOL, Outer capsid protein VP5 | | Authors: | Stuart, D.I, Sutton, G.C. | | Deposit date: | 2024-01-25 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The effect of pH on the structure of Bluetongue virus VP5.
J.Gen.Virol., 105, 2024
|
|
1J0G
 
 | | Solution Structure of Mouse Hypothetical 9.1 kDa Protein, A Ubiquitin-like Fold | | Descriptor: | Hypothetical Protein 1810045K17 | | Authors: | Zhao, C, Kigawa, T, Koshiba, S, Tochio, N, Kobayashi, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-13 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Mouse Hypothetical 9.1 kDa Protein, A Ubiquitin-like Fold
To be Published
|
|
1IRL
 
 | | THE SOLUTION STRUCTURE OF THE F42A MUTANT OF HUMAN INTERLEUKIN 2 | | Descriptor: | INTERLEUKIN-2 | | Authors: | Mott, H.R, Baines, B.S, Hall, R.M, Cooke, R.M, Driscoll, P.C, Weir, M.P, Campbell, I.D. | | Deposit date: | 1995-08-25 | | Release date: | 1995-12-07 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the F42A mutant of human interleukin 2.
J.Mol.Biol., 247, 1995
|
|
1SJU
 
 | | MINI-PROINSULIN, SINGLE CHAIN INSULIN ANALOG MUTANT: DES B30, HIS(B 10)ASP, PRO(B 28)ASP AND PEPTIDE BOND BETWEEN LYS B 29 AND GLY A 1, NMR, 20 STRUCTURES | | Descriptor: | PROINSULIN | | Authors: | Hua, Q.X, Hu, S.Q, Jia, W.H, Chu, Y.C, Burke, G.T, Wang, S.H, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 1997-10-09 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Mini-proinsulin and mini-IGF-I: homologous protein sequences encoding non-homologous structures.
J.Mol.Biol., 277, 1998
|
|
1IIO
 
 | |
4K8Y
 
 | | Atomic resolution crystal structures of Kallikrein-Related Peptidase 4 complexed with Sunflower Trypsin Inhibitor (SFTI-1) | | Descriptor: | Kallikrein-4, Trypsin inhibitor 1 | | Authors: | Ilyichova, O.V, Swedberg, J.E, de Veer, S.J, Sit, K.C, Harris, J.M, Buckle, A.M. | | Deposit date: | 2013-04-19 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Direct and indirect mechanisms of KLK4 inhibition revealed by structure and dynamics
Sci Rep, 6, 2016
|
|
4KD6
 
 | | Crystal structure of a Enoyl-CoA hydratase/isomerase from Burkholderia graminis C4D1M | | Descriptor: | Enoyl-CoA hydratase/isomerase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-24 | | Release date: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of a Enoyl-CoA hydratase/isomerase from Burkholderia graminis C4D1M
To be Published
|
|
1SJV
 
 | | Three-Dimensional Structure of a Llama VHH Domain Swapping | | Descriptor: | r9 | | Authors: | Spinelli, S, Desmyter, A, Frenken, L, Verrips, T, Cambillau, C. | | Deposit date: | 2004-03-04 | | Release date: | 2004-06-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Domain swapping of a llama VHH domain builds a crystal-wide beta-sheet structure.
Febs Lett., 564, 2004
|
|
1JAV
 
 | |
4KQE
 
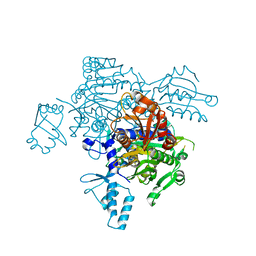 | | The mutant structure of the human glycyl-tRNA synthetase E71G | | Descriptor: | GLYCEROL, Glycine--tRNA ligase | | Authors: | Qin, X, Hao, Z, Tian, Q, Zhang, Z, Zhou, C, Xie, W. | | Deposit date: | 2013-05-15 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.739 Å) | | Cite: | Large Conformational Changes of Insertion 3 in Human Glycyl-tRNA Synthetase (hGlyRS) during Catalysis
J.Biol.Chem., 291, 2016
|
|
7PLN
 
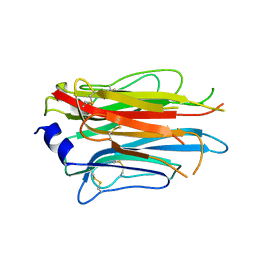 | | Structure of the APCbeta domain of Plasmodium vivax perforin-like protein 1 | | Descriptor: | Sporozoite micronemal protein essential for cell traversal, putative | | Authors: | Williams, S.I, Ni, T, Yu, X, Gilbert, R.J.C. | | Deposit date: | 2021-08-31 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural, Functional and Computational Studies of Membrane Recognition by Plasmodium Perforin-Like Proteins 1 and 2
J.Mol.Biol., 434, 2022
|
|
7RX4
 
 | | Cryo-EM reconstruction of AS2 nanotube (Form II like) | | Descriptor: | AS2 peptide | | Authors: | Wang, F, Gnewou, O.M, Solemanifar, A, Xu, C, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2021-08-21 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM of Helical Polymers.
Chem.Rev., 122, 2022
|
|
7RX5
 
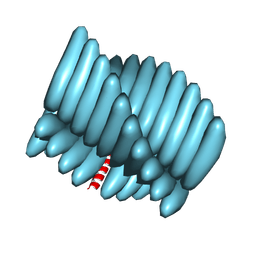 | | Cryo-EM reconstruction of Form1-N2 nanotube (Form I like) | | Descriptor: | F1-N2 nanotube | | Authors: | Wang, F, Gnewou, O.M, Solemanifar, A, Xu, C, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2021-08-21 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM of Helical Polymers.
Chem.Rev., 122, 2022
|
|
2BZG
 
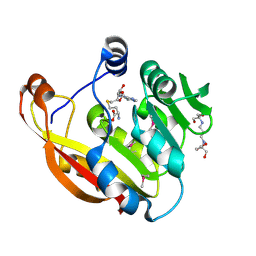 | | Crystal structure of thiopurine S-methyltransferase. | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYL-L-HOMOCYSTEINE, THIOPURINE S-METHYLTRANSFERASE | | Authors: | Battaile, K.P, Wu, H, Zeng, H, Loppnau, P, Dong, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-17 | | Release date: | 2005-08-25 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Basis of Allele Variation of Human Thiopurine-S-Methyltransferase.
Proteins, 67, 2007
|
|
2BTV
 
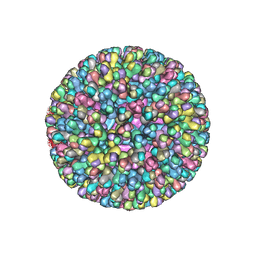 | | ATOMIC MODEL FOR BLUETONGUE VIRUS (BTV) CORE | | Descriptor: | PROTEIN (VP3 CORE PROTEIN), PROTEIN (VP7 CORE PROTEIN) | | Authors: | Grimes, J.M, Burroughs, J.N, Gouet, P, Diprose, J.M, Malby, R, Zientras, S, Mertens, P.P.C, Stuart, D.I. | | Deposit date: | 1998-09-05 | | Release date: | 1998-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The atomic structure of the bluetongue virus core.
Nature, 395, 1998
|
|
1C7M
 
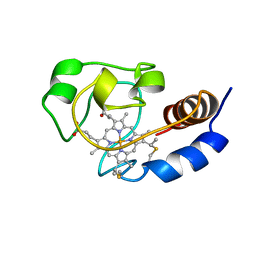 | | SOLUTION STRUCTURE OF THE FUNCTIONAL DOMAIN OF PARACOCCUS DENITRIFICANS CYTOCHROME C552 IN THE REDUCED STATE | | Descriptor: | HEME C, PROTEIN (CYTOCHROME C552) | | Authors: | Pristovsek, P, Luecke, C, Reincke, B, Ludwig, B, Rueterjans, H. | | Deposit date: | 2000-03-03 | | Release date: | 2000-07-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the functional domain of Paracoccus denitrificans cytochrome c552 in the reduced state.
Eur.J.Biochem., 267, 2000
|
|
2BOE
 
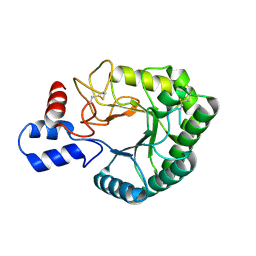 | | Catalytic domain of endo-1,4-glucanase Cel6A mutant Y73S from Thermobifida fusca | | Descriptor: | ENDOGLUCANASE E-2 | | Authors: | Larsson, A.M, Bergfors, T, Dultz, E, Irwin, D.C, Roos, A, Driguez, H, Wilson, D.B, Jones, T.A. | | Deposit date: | 2005-04-10 | | Release date: | 2005-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structure of Thermobifida Fusca Endoglucanase Cel6A in Complex with Substrate and Inhibitor: The Role of Tyrosine Y73 in Substrate Ring Distortion.
Biochemistry, 44, 2005
|
|
2BVA
 
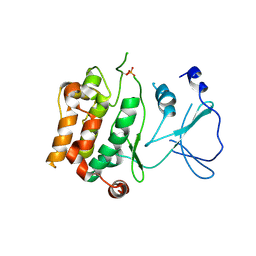 | | Crystal structure of the human P21-activated kinase 4 | | Descriptor: | P21-ACTIVATED KINASE 4 | | Authors: | Debreczeni, J.E, Bunkoczi, G, Eswaran, J, Filippakopoulos, P, Das, S, Fedorov, O, Sundstrom, M, Arrowsmith, C, Edwards, A, von Delft, F, Knapp, S. | | Deposit date: | 2005-06-23 | | Release date: | 2005-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the P21-Activated Kinases Pak4, Pak5, and Pak6 Reveal Catalytic Domain Plasticity of Active Group II Paks.
Structure, 15, 2007
|
|
2BEL
 
 | | Structure of human 11-beta-hydroxysteroid dehydrogenase in complex with NADP and carbenoxolone | | Descriptor: | CARBENOXOLONE, CHLORIDE ION, CORTICOSTEROID 11-BETA-DEHYDROGENASE ISOZYME 1, ... | | Authors: | Kavanagh, K, Wu, X, Svensson, S, Elleby, B, von Delft, F, Debreczeni, J.E, Sharma, S, Bray, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Abrahmsen, L, Oppermann, U. | | Deposit date: | 2004-11-25 | | Release date: | 2004-12-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The High Resolution Structures of Human, Murine and Guinea Pig 11-Beta-Hydroxysteroid Dehydrogenase Type 1 Reveal Critical Differences in Active Site Architecture
To be Published
|
|
8SUE
 
 | | Human asparagine synthetase (apo-ASNS) | | Descriptor: | Asparagine synthetase [glutamine-hydrolyzing] | | Authors: | Coricello, A, Zhu, W, Lupia, A, Gratteri, C, Vos, M, Chaptal, V, Alcaro, S, Takagi, Y, Richards, N. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Human asparagine synthetase (apo-ASNS)
To Be Published
|
|
