7PBW
 
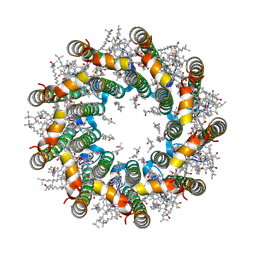 | | Cryo-EM structure of light harvesting complex 2 from Rba. sphaeroides. | | Descriptor: | BACTERIOCHLOROPHYLL A, CALCIUM ION, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Qian, P, Swainsbury, D.J.K, Croll, T.I, Castro-Hartmann, P, Sader, K, Divitini, G, Hunter, C.N. | | Deposit date: | 2021-08-02 | | Release date: | 2021-11-24 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Cryo-EM Structure of the Rhodobacter sphaeroides Light-Harvesting 2 Complex at 2.1 angstrom.
Biochemistry, 60, 2021
|
|
3HQ5
 
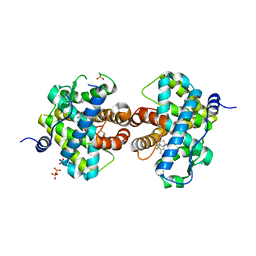 | | Progesterone Receptor bound to an Alkylpyrrolidine ligand. | | Descriptor: | 2-chloro-4-{[(3S)-1-methylpyrrolidin-3-yl][2-(trifluoromethyl)benzyl]amino}benzonitrile, GLYCEROL, Progesterone receptor, ... | | Authors: | Madauss, K.P, Williams, S.P, Washburn, D.G. | | Deposit date: | 2009-06-05 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational design of orally-active, pyrrolidine-based progesterone receptor partial agonists.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6DSL
 
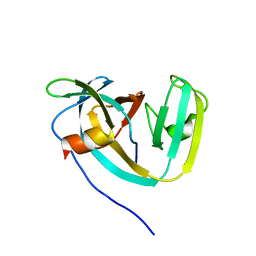 | | Consensus engineered intein (Cat) with atypical split site | | Descriptor: | Consensus engineered intein CatC, Consensus engineered intein CatN | | Authors: | Sekar, G, Stevens, A.J, Muir, T.W, Cowburn, D. | | Deposit date: | 2018-06-14 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | An Atypical Mechanism of Split Intein Molecular Recognition and Folding.
J. Am. Chem. Soc., 140, 2018
|
|
3EAC
 
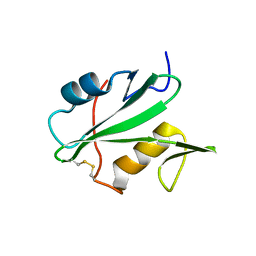 | |
3G8O
 
 | | Progesterone Receptor with bound Pyrrolidine 1 | | Descriptor: | N~2~-[4-cyano-3-(trifluoromethyl)phenyl]-N,N-dimethyl-N~2~-(2,2,2-trifluoroethyl)-L-alaninamide, Progesterone receptor, SULFATE ION | | Authors: | Thompson, S.K, Washburn, D.G, Madauss, K.P, Williams, S.P, Stewart, E.L. | | Deposit date: | 2009-02-12 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational design of orally-active, pyrrolidine-based progesterone receptor partial agonists.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3H52
 
 | | Crystal structure of the antagonist form of human glucocorticoid receptor | | Descriptor: | 11-(4-DIMETHYLAMINO-PHENYL)-17-HYDROXY-13-METHYL-17-PROP-1-YNYL-1,2,6,7,8,11,12,13,14,15,16,17-DODEC AHYDRO-CYCLOPENTA[A]PHENANTHREN-3-ONE, GLYCEROL, Glucocorticoid receptor, ... | | Authors: | Schoch, G.A, Benz, J, D'Arcy, B, Stihle, M, Burger, D, Thoma, R, Ruf, A. | | Deposit date: | 2009-04-21 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular switch in the glucocorticoid receptor: active and passive antagonist conformations
J.Mol.Biol., 395, 2010
|
|
1AB2
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE SRC HOMOLOGY 2 DOMAIN OF C-ABL | | Descriptor: | C-ABL TYROSINE KINASE SH2 DOMAIN | | Authors: | Overduin, M, Rios, C.B, Mayer, B.J, Baltimore, D, Cowburn, D. | | Deposit date: | 1993-07-19 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the src homology 2 domain of c-abl.
Cell(Cambridge,Mass.), 70, 1992
|
|
3INU
 
 | | Crystal structure of an unbound KZ52 neutralizing anti-Ebolavirus antibody. | | Descriptor: | GLYCEROL, KZ52 antibody fragment heavy chain, KZ52 antibody fragment light chain, ... | | Authors: | Lee, J.E, Fusco, M.L, Abelson, D.M, Hessell, A.J, Burton, D.R, Saphire, E.O. | | Deposit date: | 2009-08-12 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Techniques and tactics used in determining the structure of the trimeric ebolavirus glycoprotein.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
7AZ7
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 37 bound | | Descriptor: | Beta sliding clamp, FORMIC ACID, PENTAETHYLENE GLYCOL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZ6
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 36 bound | | Descriptor: | ACETATE ION, Beta sliding clamp, CHLORIDE ION, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
1BZV
 
 | | [D-ALAB26]-DES(B27-B30)-INSULIN-B26-AMIDE A SUPERPOTENT SINGLE-REPLACEMENT INSULIN ANALOGUE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | INSULIN | | Authors: | Kurapkat, G, Siedentopf, M, Gattner, H.G, Hagelstein, M, Brandenburg, D, Grotzinger, J, Wollmer, A. | | Deposit date: | 1998-11-04 | | Release date: | 1999-05-18 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a superpotent B-chain-shortened single-replacement insulin analogue.
Protein Sci., 8, 1999
|
|
4UUJ
 
 | | POTASSIUM CHANNEL KCSA-FAB WITH TETRAHEXYLAMMONIUM | | Descriptor: | ANTIBODY FAB FRAGMENT HEAVY CHAIN, ANTIBODY FAB FRAGMENT LIGHT CHAIN, COBALT (II) ION, ... | | Authors: | Lenaeus, M.J, Burdette, D, Wagner, T, Focia, P.J, Gross, A. | | Deposit date: | 2014-07-29 | | Release date: | 2014-08-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Kcsa in Complex with Symmetrical Quaternary Ammonium Compounds Reveal a Hydrophobic Binding Site.
Biochemistry, 53, 2014
|
|
2LMB
 
 | | Solution Structure of C-terminal RAGE (ctRAGE) | | Descriptor: | Advanced glycosylation end product-specific receptor | | Authors: | Rai, V, Maldonado, A.Y, Burz, D.S, Reverdatto, S, Schmidt, A, Shekhtman, A. | | Deposit date: | 2011-11-29 | | Release date: | 2011-12-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Signal transduction in receptor for advanced glycation end products (RAGE): solution structure of C-terminal rage (ctRAGE) and its binding to mDia1.
J.Biol.Chem., 287, 2012
|
|
3HI1
 
 | | Structure of HIV-1 gp120 (core with V3) in Complex with CD4-Binding-Site Antibody F105 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F105 Heavy Chain, F105 Light Chain, ... | | Authors: | Kwon, Y.D, Chen, L, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z, Zhang, M.-Y, Arthos, J, Burton, D.R, Dimitrov, D, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-05-18 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
3IDX
 
 | | Crystal structure of HIV-gp120 core in complex with CD4-binding site antibody b13, space group C222 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fab b13 heavy chain, ... | | Authors: | Chen, L, Kwon, Y.D, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z.Y, Zhang, M.Y, Arthos, J, Burton, D.R, Dimitrov, D.S, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-07-22 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
7SGT
 
 | |
3IDY
 
 | | Crystal structure of HIV-gp120 core in complex with CD4-binding site antibody b13, space group C2221 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab b13 heavy chain, Fab b13 light chain, ... | | Authors: | Chen, L, Kwon, Y.D, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z.Y, Zhang, M.Y, Arthos, J, Burton, D.R, Dimitrov, D.S, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-07-22 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
1BAK
 
 | |
1CU4
 
 | | CRYSTAL STRUCTURE OF THE ANTI-PRION FAB 3F4 IN COMPLEX WITH ITS PEPTIDE EPITOPE | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, RECOGNITION PEPTIDE | | Authors: | Kanyo, Z.F, Pan, K.M, Williamson, R.A, Burton, D.R, Prusiner, S.B, Fletterick, R.J, Cohen, F.E. | | Deposit date: | 1999-08-20 | | Release date: | 2000-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Antibody binding defines a structure for an epitope that participates in the PrPC-->PrPSc conformational change.
J.Mol.Biol., 293, 1999
|
|
8PAY
 
 | | Structure of the E.coli DNA polymerase sliding clamp with a covalently bound peptide 2. | | Descriptor: | ACE-GLN-ALC-GLC-LEU-PHE, Beta sliding clamp, GLYCEROL, ... | | Authors: | Compain, G, Monsarrat, C, Blagojevic, J, Brillet, K, Dumas, P, Hammann, P, Kuhn, L, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Burnouf, D, wagner, J, Guichard, G. | | Deposit date: | 2023-06-08 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Peptide-Based Covalent Inhibitors Bearing Mild Electrophiles to Target a Conserved His Residue of the Bacterial Sliding Clamp.
Jacs Au, 4, 2024
|
|
8PAT
 
 | | Structure of the E.coli DNA polymerase sliding clamp with a covalently bound peptide 3. | | Descriptor: | ACE-GLN-ALC-GLX-LEU-PHE, Beta sliding clamp | | Authors: | Compain, G, Monsarrat, C, Blagojevic, J, Brillet, K, Dumas, P, Hammann, P, Kuhn, L, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Burnouf, D, Guichard, G. | | Deposit date: | 2023-06-08 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Peptide-Based Covalent Inhibitors Bearing Mild Electrophiles to Target a Conserved His Residue of the Bacterial Sliding Clamp.
Jacs Au, 4, 2024
|
|
1CR9
 
 | | CRYSTAL STRUCTURE OF THE ANTI-PRION FAB 3F4 | | Descriptor: | FAB ANTIBODY HEAVY CHAIN, FAB ANTIBODY LIGHT CHAIN | | Authors: | Kanyo, Z.F, Pan, K.M, Williamson, R.A, Burton, D.R, Prusiner, S.B, Fletterick, R.J, Cohen, F.E. | | Deposit date: | 1999-08-14 | | Release date: | 2000-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antibody binding defines a structure for an epitope that participates in the PrPC-->PrPSc conformational change.
J.Mol.Biol., 293, 1999
|
|
1Z66
 
 | | NMR solution structure of domain III of E-protein of tick-borne Langat flavivirus (no RDC restraints) | | Descriptor: | Major envelope protein E | | Authors: | Mukherjee, M, Dutta, K, White, M.A, Cowburn, D, Fox, R.O. | | Deposit date: | 2005-03-21 | | Release date: | 2006-03-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and backbone dynamics of domain III of the E protein of tick-borne Langat flavivirus suggests a potential site for molecular recognition.
Protein Sci., 15, 2006
|
|
1BDB
 
 | | CIS-BIPHENYL-2,3-DIHYDRODIOL-2,3-DEHYDROGENASE FROM PSEUDOMONAS SP. LB400 | | Descriptor: | CIS-BIPHENYL-2,3-DIHYDRODIOL-2,3-DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Huelsmeyer, M, Hecht, H.-J, Niefind, K, Hofer, B, Timmis, K.N, Schomburg, D. | | Deposit date: | 1997-05-10 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of cis-biphenyl-2,3-dihydrodiol-2,3-dehydrogenase from a PCB degrader at 2.0 A resolution.
Protein Sci., 7, 1998
|
|
1HPT
 
 | |
