2HKF
 
 | | Crystal structure of the Complex Fab M75- Peptide | | Descriptor: | Carbonic anhydrase 9, Immunoglobulin Heavy chain Fab fragment, Immunoglobulin Light chain Fab fragment | | Authors: | Kral, V, Mader, P, Stouracova, R, Fabry, M, Horejsi, M, Brynda, J. | | Deposit date: | 2006-07-04 | | Release date: | 2007-11-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Stabilization of antibody structure upon association to a human carbonic anhydrase IX epitope studied by X-ray crystallography, microcalorimetry, and molecular dynamics simulations.
Proteins, 71, 2008
|
|
8CC2
 
 | | Cathepsin B1 from Schistosoma mansoni in complex with gallinamide A | | Descriptor: | Cathepsin B-like peptidase (C01 family), SODIUM ION, gallinamide A, ... | | Authors: | Rubesova, P, Brynda, J, Mares, M, Gerwick, W.H. | | Deposit date: | 2023-01-26 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Cathepsin B1 from Schistosoma mansoni in complex with gallinamide A
To Be Published
|
|
8CCU
 
 | | Cathepsin B1 from Schistosoma mansoni in complex with gallinamide analog 1 | | Descriptor: | Cathepsin B-like peptidase (C01 family), SODIUM ION, [(2~{S})-1-[[(2~{S})-1-[[(2~{S})-5-[(2~{S})-3-methoxy-2-(2-methylpropyl)-5-oxidanylidene-2~{H}-pyrrol-1-yl]-5-oxidanylidene-pentan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl] (2~{S},3~{S})-2-(dimethylamino)-3-methyl-pentanoate | | Authors: | Rubesova, P, Brynda, J, Gerwick, W.H, Mares, M. | | Deposit date: | 2023-01-27 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cathepsin B1 from Schistosoma mansoni in complex with gallinamide analog 1
To Be Published
|
|
1JP5
 
 | | Crystal structure of the single-chain Fv fragment 1696 in complex with the epitope peptide corresponding to N-terminus of HIV-1 protease | | Descriptor: | epitope peptide corresponding to N-terminus of HIV-1 protease, single-chain Fv fragment 1696 | | Authors: | Rezacova, P, Lescar, J, Brynda, J, Fabry, M, Horejsi, M, Sedlacek, J, Bentley, G.A. | | Deposit date: | 2001-08-01 | | Release date: | 2001-10-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of HIV-1 and HIV-2 protease inhibition by a monoclonal antibody.
Structure, 9, 2001
|
|
1SP5
 
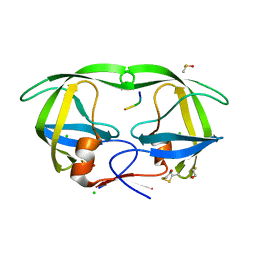 | | Crystal structure of HIV-1 protease complexed with a product of autoproteolysis | | Descriptor: | 5-mer peptide from Protease, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Vondrackova, E, Hasek, J, Jaskolski, M, Rezacova, P, Dohnalek, J, Skalova, T, Petrokova, H, Duskova, J, Brynda, J, Sedlacek, J. | | Deposit date: | 2004-03-16 | | Release date: | 2005-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Product of enzymatic self-cleavage bound in the active site of HIV protease
To be Published
|
|
7AS0
 
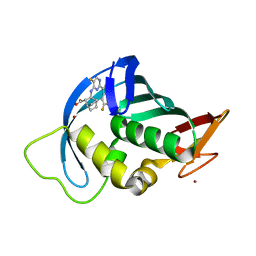 | | Influenza A PB2 in complex with VX-787 | | Descriptor: | (2S,3S)-3-[[5-fluoranyl-2-(5-fluoranyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, BROMIDE ION, Polymerase basic protein 2 | | Authors: | Radilova, K, Brynda, J. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Thermodynamic Analysis of the Resistance Development to Pimodivir (VX-787), the Clinical Inhibitor of Cap Binding to PB2 Subunit of Influenza A Polymerase.
Molecules, 26, 2021
|
|
7AS1
 
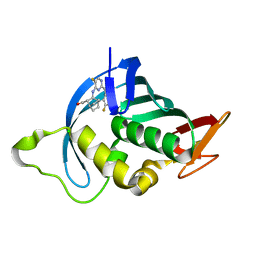 | | Influenza A PB2 (F404Y mutation) in complex with VX-787 | | Descriptor: | (2S,3S)-3-[[5-fluoranyl-2-(5-fluoranyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, CHLORIDE ION, Polymerase basic protein 2 | | Authors: | Radilova, K, Brynda, J. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Thermodynamic Analysis of the Resistance Development to Pimodivir (VX-787), the Clinical Inhibitor of Cap Binding to PB2 Subunit of Influenza A Polymerase.
Molecules, 26, 2021
|
|
7AS2
 
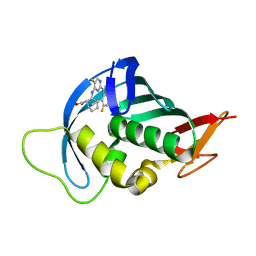 | | Influenza A PB2 (M431 mutation) in complex with VX-787 | | Descriptor: | (2S,3S)-3-[[5-fluoranyl-2-(5-fluoranyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, Polymerase basic protein 2 | | Authors: | Radilova, K, Brynda, J. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Thermodynamic Analysis of the Resistance Development to Pimodivir (VX-787), the Clinical Inhibitor of Cap Binding to PB2 Subunit of Influenza A Polymerase.
Molecules, 26, 2021
|
|
7AS3
 
 | | Influenza A PB2 (H357N mutation) in complex with VX-787 | | Descriptor: | (2S,3S)-3-[[5-fluoranyl-2-(5-fluoranyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, Polymerase basic protein 2 | | Authors: | Radilova, K, Brynda, J. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Thermodynamic Analysis of the Resistance Development to Pimodivir (VX-787), the Clinical Inhibitor of Cap Binding to PB2 Subunit of Influenza A Polymerase.
Molecules, 26, 2021
|
|
6S03
 
 | | Carbonic Anhydrase CAIX mimic in complex with inhibitor I39LT379 | | Descriptor: | 4-[[4-[5,5-dimethyl-2-(6-methylpyridin-2-yl)-4,6-dihydropyrrolo[1,2-b]pyrazol-3-yl]pyridin-2-yl]amino]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Kugler, M, Brynda, J, Rezacova, P. | | Deposit date: | 2019-06-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Identification of Novel Carbonic Anhydrase IX Inhibitors Using High-Throughput Screening of Pooled Compound Libraries by DNA-Linked Inhibitor Antibody Assay (DIANA).
SLAS Discov, 25, 2020
|
|
6RZX
 
 | | Carbonic Anhydrase CAIX mimic in complex with inhibitor FBSA | | Descriptor: | 1,1,2,2,3,3,4,4,4-nonakis(fluoranyl)butane-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Kugler, M, Brynda, J, Rezacova, P. | | Deposit date: | 2019-06-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Identification of Novel Carbonic Anhydrase IX Inhibitors Using High-Throughput Screening of Pooled Compound Libraries by DNA-Linked Inhibitor Antibody Assay (DIANA).
SLAS Discov, 25, 2020
|
|
8A2K
 
 | | human STING in complex with 2'-3'-cyclic-GMP-7-deaza(4-[(2-naphthyloxy)methyl]phenyl)-AMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{S},15~{R},17~{R},18~{R})-8-[4-azanyl-5-[4-(naphthalen-1-yloxymethyl)phenyl]pyrrolo[2,3-d]pyrimidin-7-yl]-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-3~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Vavrina, Z, Brynda, J, Rezacova, P. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Design, Synthesis, and Biochemical and Biological Evaluation of Novel 7-Deazapurine Cyclic Dinucleotide Analogues as STING Receptor Agonists.
J.Med.Chem., 65, 2022
|
|
8A2H
 
 | | human STING in complex with 2',3'-cyclic-GMP-7-deazaphenyl-AMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{S},15~{R},17~{R},18~{R})-8-(4-azanyl-5-phenyl-pyrrolo[2,3-d]pyrimidin-7-yl)-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Smola, M, Vavrina, Z, Boura, E, Brynda, J. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design, Synthesis, and Biochemical and Biological Evaluation of Novel 7-Deazapurine Cyclic Dinucleotide Analogues as STING Receptor Agonists.
J.Med.Chem., 65, 2022
|
|
8A2J
 
 | | human STING in complex with 2'-3'-cyclic-GMP-7-deaza(4-biphenylyl)-AMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{S},15~{R},17~{R},18~{R})-8-[4-azanyl-5-(4-phenylphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Vavrina, Z, Brynda, J, Rezacova, P. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Design, Synthesis, and Biochemical and Biological Evaluation of Novel 7-Deazapurine Cyclic Dinucleotide Analogues as STING Receptor Agonists.
J.Med.Chem., 65, 2022
|
|
8A2I
 
 | | human STING in complex with 2'-3'-cyclic-GMP-7-deaza(4-(2-naphthyl)phenyl)-AMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{S},15~{R},17~{R},18~{R})-8-[4-azanyl-5-(4-naphthalen-2-ylphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-3~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Vavrina, Z, Brynda, J, Rezacova, P. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Design, Synthesis, and Biochemical and Biological Evaluation of Novel 7-Deazapurine Cyclic Dinucleotide Analogues as STING Receptor Agonists.
J.Med.Chem., 65, 2022
|
|
1RL8
 
 | | Crystal structure of the complex of resistant strain of hiv-1 protease(v82a mutant) with ritonavir | | Descriptor: | RITONAVIR, protease RETROPEPSIN | | Authors: | Rezacova, P, Brynda, J, Sedlacek, J, Konvalinka, J, Fabry, M, Horejsi, M. | | Deposit date: | 2003-11-25 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex of resistant strain of hiv-1 protease(v82a mutant) with ritonavir
To be Published, 2005
|
|
1SVZ
 
 | | Crystal structure of the single-chain Fv fragment 1696 in complex with the epitope peptide corresponding to N-terminus of HIV-2 protease | | Descriptor: | epitope peptide corresponding to N-terminus of HIV-2 protease, single-chain Fv fragment 1696 | | Authors: | Rezacova, P, Brynda, J, Lescar, J, Bentley, G.A, Fabry, M, Horejsi, M, Sedlacek, J. | | Deposit date: | 2004-03-30 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of a cross-reaction complex between an anti-HIV-1 protease antibody and an HIV-2 protease peptide
J.Struct.Biol., 149, 2005
|
|
4O1G
 
 | | MTB adenosine kinase in complex with gamma-Thio-ATP | | Descriptor: | Adenosine kinase, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, SODIUM ION | | Authors: | Dostal, J, Brynda, J, Hocek, M, Pichova, I. | | Deposit date: | 2013-12-15 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Inhibition of Mycobacterial and Human Adenosine Kinase by 7-Substituted 7-(Het)aryl-7-deazaadenine Ribonucleosides
J.Med.Chem., 57, 2014
|
|
7AGE
 
 | |
7AGB
 
 | | Protease Sapp1p from Candida parapsilosis in complex with KB70 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Candidapepsin, ... | | Authors: | Dostal, J, Heidingsfeld, O, Brynda, J. | | Deposit date: | 2020-09-22 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural determinants for subnanomolar inhibition of the secreted aspartic protease Sapp1p from Candida parapsilosis .
J Enzyme Inhib Med Chem, 36, 2021
|
|
7AGD
 
 | | Protease Sapp1p from Candida parapsilosis in complex with KB75 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Candidapepsin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dostal, J, Heidingsfeld, O, Brynda, J. | | Deposit date: | 2020-09-22 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural determinants for subnanomolar inhibition of the secreted aspartic protease Sapp1p from Candida parapsilosis .
J Enzyme Inhib Med Chem, 36, 2021
|
|
7AGC
 
 | | Protease Sapp1p from Candida parapsilosis in complex with KB74 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Candidapepsin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dostal, J, Heidingsfeld, O, Brynda, J. | | Deposit date: | 2020-09-22 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural determinants for subnanomolar inhibition of the secreted aspartic protease Sapp1p from Candida parapsilosis .
J Enzyme Inhib Med Chem, 36, 2021
|
|
3V7X
 
 | | Complex of human carbonic anhydrase II with N-[2-(3,4-dimethoxyphenyl)ethyl]-4-sulfamoylbenzamide | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, MERCURIBENZOIC ACID, ... | | Authors: | Mader, P, Brynda, J, Rezacova, P. | | Deposit date: | 2011-12-22 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Synthesis, Structure-Activity Relationship Studies, and X-ray Crystallographic Analysis of Arylsulfonamides as Potent Carbonic Anhydrase Inhibitors.
J.Med.Chem., 55, 2012
|
|
3VBD
 
 | | Complex of human carbonic anhydrase II with 4-(6-methoxy-3,4-dihydroisoquinolin-1-yl)benzenesulfonamide | | Descriptor: | 4-(6-methoxy-3,4-dihydroisoquinolin-1-yl)benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Mader, P, Brynda, J, Rezacova, P. | | Deposit date: | 2012-01-02 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Synthesis, Structure-Activity Relationship Studies, and X-ray Crystallographic Analysis of Arylsulfonamides as Potent Carbonic Anhydrase Inhibitors.
J.Med.Chem., 55, 2012
|
|
3IGP
 
 | | Structure of inhibitor binding to Carbonic Anhydrase II | | Descriptor: | 6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Gitto, R, Agnello, S, Brynda, J, Mader, P, Supuran, C.T, Chimirri, A. | | Deposit date: | 2009-07-28 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of 3,4-Dihydroisoquinoline-2(1H)-sulfonamides as Potent Carbonic Anhydrase Inhibitors: Synthesis, Biological Evaluation, and Enzyme-Ligand X-ray Studies.
J.Med.Chem., 2010
|
|
