1ZKZ
 
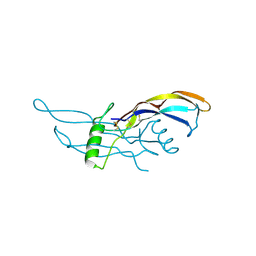 | | Crystal Structure of BMP9 | | Descriptor: | Growth/differentiation factor 2 | | Authors: | Brown, M.A, Zhao, Q, Baker, K.A, Naik, C, Chen, C, Pukac, L, Singh, M, Tsareva, T, Parice, Y, Mahoney, A, Roschke, V, Sanyal, I, Choe, S. | | Deposit date: | 2005-05-04 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of BMP-9 and functional interactions with pro-region and receptors
J.Biol.Chem., 280, 2005
|
|
3U1Y
 
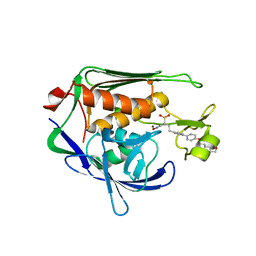 | | Potent Inhibitors of LpxC for the Treatment of Gram-Negative Infections | | Descriptor: | (2R)-N-hydroxy-2-methyl-2-(methylsulfonyl)-4-{4'-[3-(morpholin-4-yl)propoxy]biphenyl-4-yl}butanamide, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Brown, M, Abramite, J, Liu, S. | | Deposit date: | 2011-09-30 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent inhibitors of LpxC for the treatment of Gram-negative infections.
J.Med.Chem., 55, 2012
|
|
4G2C
 
 | | DyP2 from Amycolatopsis sp. ATCC 39116 | | Descriptor: | ACETATE ION, DyP2, MANGANESE (II) ION, ... | | Authors: | Brown, M.E, Barros, T, Chang, M.C.Y. | | Deposit date: | 2012-07-11 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification and characterization of a multifunctional dye peroxidase from a lignin-reactive bacterium.
Acs Chem.Biol., 7, 2012
|
|
3NYA
 
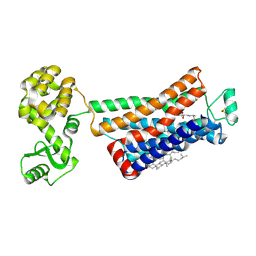 | | Crystal structure of the human beta2 adrenergic receptor in complex with the neutral antagonist alprenolol | | Descriptor: | (2S)-1-[(1-methylethyl)amino]-3-(2-prop-2-en-1-ylphenoxy)propan-2-ol, Beta-2 adrenergic receptor, Lysozyme, ... | | Authors: | Brown, M.A, Wacker, D, Fenalti, G, Katritch, V, Abagyan, R, Cherezov, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Conserved Binding Mode of Human beta(2) Adrenergic Receptor Inverse Agonists and Antagonist Revealed by X-ray Crystallography
J.Am.Chem.Soc., 132, 2010
|
|
1MMQ
 
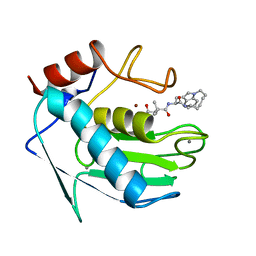 | | MATRILYSIN COMPLEXED WITH HYDROXAMATE INHIBITOR | | Descriptor: | CALCIUM ION, MATRILYSIN, N4-HYDROXY-2-ISOBUTYL-N1-(9-OXO-1,8-DIAZA-TRICYCLO[10.6.1.013,18]NONADECA-12(19),13,15,17-TETRAEN-10-YL)-SUCCINAMIDE, ... | | Authors: | Browner, M.F, Smith, W.W, Castelhano, A.L. | | Deposit date: | 1995-03-22 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Matrilysin-inhibitor complexes: common themes among metalloproteases.
Biochemistry, 34, 1995
|
|
1MMP
 
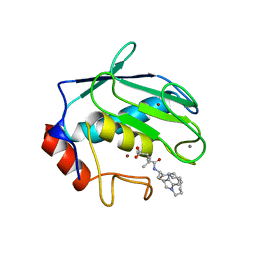 | | MATRILYSIN COMPLEXED WITH CARBOXYLATE INHIBITOR | | Descriptor: | 5-METHYL-3-(9-OXO-1,8-DIAZA-TRICYCLO[10.6.1.013,18]NONADECA-12(19),13,15,17-TETRAEN-10-YLCARBAMOYL)-HEXANOIC ACID, CALCIUM ION, GELATINASE A, ... | | Authors: | Browner, M.F, Smith, W.W, Castelhano, A.L. | | Deposit date: | 1995-03-22 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Matrilysin-inhibitor complexes: common themes among metalloproteases.
Biochemistry, 34, 1995
|
|
1MMR
 
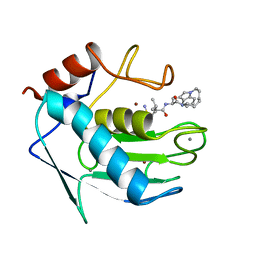 | | MATRILYSIN COMPLEXED WITH SULFODIIMINE INHIBITOR | | Descriptor: | 4-METHYL-3-(9-OXO-1,8-DIAZA-TRICYCLO[10.6.1.0(13,18)]NONADECA-12(19),13(18),15,17-TETRAENE-10-CARBAMOYL)PENTA-METHYLSULFONEDIIMINE, CALCIUM ION, MATRILYSIN, ... | | Authors: | Browner, M.F, Smith, W.W, Castelhano, A.L. | | Deposit date: | 1995-03-22 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Matrilysin-inhibitor complexes: common themes among metalloproteases.
Biochemistry, 34, 1995
|
|
1MS6
 
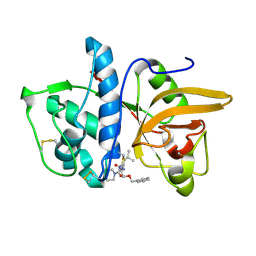 | | Dipeptide Nitrile Inhibitor Bound to Cathepsin S. | | Descriptor: | Cathepsin S, MORPHOLINE-4-CARBOXYLIC ACID [1S-(2-BENZYLOXY-1R-CYANO-ETHYLCARBAMOYL)-3-METHYL-BUTYL]AMIDE | | Authors: | Ward, Y.D, Thomson, D.S, Frye, L.L, Cywin, C.L, Morwick, T, Emmanuel, M.J, Zindell, R, McNeil, D, Bekkali, Y, Giradot, M, Hrapchak, M, DeTuri, M, Crane, K, White, D, Pav, S, Wang, Y, Hao, M.H, Grygon, C.A, Labadia, M.E, Freeman, D.M, Davidson, W, Hopkins, J.L, Brown, M.L, Spero, D.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-04-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and synthesis of dipeptide nitriles as reversible and potent Cathepsin S inhibitors
J.Med.Chem., 45, 2002
|
|
6RB9
 
 | | The pore structure of Clostridium perfringens epsilon toxin | | Descriptor: | Epsilon-toxin type B | | Authors: | Savva, C.G, Clark, A.R, Naylor, C.E, Popoff, M.R, Moss, D.S, Basak, A.K, Titball, R.W, Bokori-Brown, M. | | Deposit date: | 2019-04-09 | | Release date: | 2019-06-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The pore structure of Clostridium perfringens epsilon toxin.
Nat Commun, 10, 2019
|
|
1VZO
 
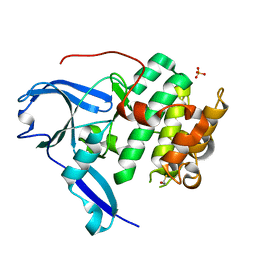 | | The structure of the N-terminal kinase domain of MSK1 reveals a novel autoinhibitory conformation for a dual kinase protein | | Descriptor: | BETA-MERCAPTOETHANOL, RIBOSOMAL PROTEIN S6 KINASE ALPHA 5, SULFATE ION | | Authors: | Smith, K.J, Carter, P.S, Bridges, A, Horrocks, P, Lewis, C, Pettman, G, Clarke, A, Brown, M, Hughes, J, Wilkinson, M, Bax, B, Reith, A. | | Deposit date: | 2004-05-21 | | Release date: | 2004-06-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of Msk1 Reveals a Novel Autoinhibitory Conformation for a Dual Kinase Protein
Structure, 12, 2004
|
|
2R9M
 
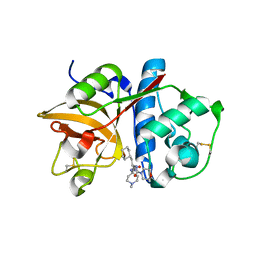 | | Cathepsin S complexed with Compound 15 | | Descriptor: | Cathepsin S, N-[(1S)-2-[(4-cyano-1-methylpiperidin-4-yl)amino]-1-(cyclohexylmethyl)-2-oxoethyl]morpholine-4-carboxamide | | Authors: | Ward, Y.D, Emmanuel, M.J, Thomson, D.S, Liu, W, Bekkali, Y, Frye, L.L, Girardot, M, Morwick, T, Young, E.R.R, Zindell, R, Hrapchak, M, DeTuri, M, White, A, Crane, K.M, White, D.M, Wang, Y, Hao, M.-H, Grygon, C.A, Labadia, M.E, Wildeson, J, Freeman, D, Nelson, R, Capolino, A, Peterson, J.D, Raymond, E.L, Brown, M.L, Spero, D.M. | | Deposit date: | 2007-09-13 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Design and Synthesis of Reversible Inhibitors of Cathepsin S: alpha,alpha-Disubstitution at the P1 Residue Provides Potent Inhibitors in Cellular Assays and In Vivo Models of Antigen Presentation
To be Published
|
|
2R9O
 
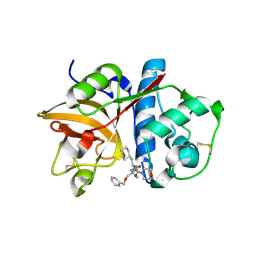 | | Cathepsin S complexed with Compound 8 | | Descriptor: | Cathepsin S, N-[(1S)-2-{[(1R)-2-(benzyloxy)-1-cyano-1-methylethyl]amino}-1-(cyclohexylmethyl)-2-oxoethyl]morpholine-4-carboxamide | | Authors: | Ward, Y.D, Emmanuel, M.J, Thomson, D.S, Liu, W, Bekkali, Y, Frye, L.L, Girardot, M, Morwick, T, Young, E.R.R, Zindell, R, Hrapchak, M, DeTuri, M, White, A, Crane, K.M, White, D.M, Wang, Y, Hao, M.-H, Grygon, C.A, Labadia, M.E, Wildeson, J, Freeman, D, Nelson, R, Capolino, A, Peterson, J.D, Raymond, E.L, Brown, M.L, Spero, D.M. | | Deposit date: | 2007-09-13 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Reversible Inhibitors of Cathepsin S: alpha,alpha-Disubstitution at the P1 Residue Provides Potent Inhibitors in Cellular Assays and In Vivo Models of Antigen Presentation
to be published
|
|
2R9N
 
 | | Cathepsin S complexed with Compound 26 | | Descriptor: | Cathepsin S, N-[(1S)-2-{[(3S)-1-benzyl-3-cyanopyrrolidin-3-yl]amino}-1-(cyclohexylmethyl)-2-oxoethyl]morpholine-4-carboxamide | | Authors: | Ward, Y.D, Emmanuel, M.J, Thomson, D.S, Liu, W, Bekkali, Y, Frye, L.L, Girardot, M, Morwick, T, Young, E.R.R, Zindell, R, Hrapchak, M, DeTuri, M, White, A, Crane, K.M, White, D.M, Wang, Y, Hao, M.-H, Grygon, C.A, Labadia, M.E, Wildeson, J, Freeman, D, Nelson, R, Capolino, A, Peterson, J.D, Raymond, E.L, Brown, M.L, Spero, D.M. | | Deposit date: | 2007-09-13 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Reversible Inhibitors of Cathepsin S: alpha,alpha-Disubstitution at the P1 Residue Provides Potent Inhibitors in Cellular Assays and In Vivo Models of Antigen Presentation
to be published
|
|
5A2F
 
 | | Two membrane distal IgSF domains of CD166 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CD166 ANTIGEN | | Authors: | Chappell, P.E, Johnson, S, Lea, S.M, Brown, M.H. | | Deposit date: | 2015-05-18 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structures of Cd6 and its Ligand Cd166 Give Insight Into Their Interaction.
Structure, 23, 2015
|
|
3ZJX
 
 | | Clostridium perfringens epsilon toxin mutant H149A bound to octyl glucoside | | Descriptor: | EPSILON-TOXIN, PHOSPHATE ION, octyl beta-D-glucopyranoside | | Authors: | Bokori-Brown, M, Kokkinidou, M.C, Savva, C.G, Fernandes da Costa, S.P, Naylor, C.E, Cole, A.R, Basak, A.K, Titball, R.W. | | Deposit date: | 2013-01-20 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Clostridium Perfringens Epsilon Toxin H149A Mutant as a Platform for Receptor Binding Studies.
Protein Sci., 22, 2013
|
|
6YBA
 
 | | HAdV-F41 Capsid | | Descriptor: | Hexon protein, Hexon-interlacing protein, Penton protein, ... | | Authors: | Perez Illana, M, Martinez, M, Mangroo, C, Brown, M, Marabini, R, San Martin, C. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-10 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of enteric adenovirus HAdV-F41 highlights structural variations among human adenoviruses.
Sci Adv, 7, 2021
|
|
4H56
 
 | | Crystal structure of the Clostridium perfringens NetB toxin in the membrane inserted form | | Descriptor: | Necrotic enteritis toxin B | | Authors: | Savva, C.G, Fernandes da Costa, S.P, Bokori-Brown, M, Naylor, C, Cole, A.R, Moss, D.S, Titball, R.W, Basak, A.K. | | Deposit date: | 2012-09-18 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Molecular Architecture and Functional Analysis of NetB, a Pore-forming Toxin from Clostridium perfringens.
J.Biol.Chem., 288, 2013
|
|
1NAU
 
 | | NMR Solution Structure of the Glucagon Antagonist [desHis1, desPhe6, Glu9]Glucagon Amide in the Presence of Perdeuterated Dodecylphosphocholine Micelles | | Descriptor: | Glucagon | | Authors: | Ying, J, Ahn, J.-M, Jacobsen, N.E, Brown, M.F, Hruby, V.J. | | Deposit date: | 2002-11-28 | | Release date: | 2003-03-18 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Glucagon Antagonist [desHis1, desPhe6, Glu9]Glucagon Amide in the Presence of Perdeuterated Dodecylphosphocholine Micelles
Biochemistry, 42, 2003
|
|
1Y6A
 
 | | Crystal structure of VEGFR2 in complex with a 2-anilino-5-aryl-oxazole inhibitor | | Descriptor: | N-[5-(ETHYLSULFONYL)-2-METHOXYPHENYL]-5-[3-(2-PYRIDINYL)PHENYL]-1,3-OXAZOL-2-AMINE, Vascular endothelial growth factor receptor 2 | | Authors: | Harris, P.A, Cheung, M, Hunter, R.N, Brown, M.L, Veal, J.M, Nolte, R.T, Wang, L, Liu, W, Crosby, R.M, Johnson, J.H, Epperly, A.H, Kumar, R, Luttrell, D.K, Stafford, J.A. | | Deposit date: | 2004-12-05 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and evaluation of 2-anilino-5-aryloxazoles as a novel class of VEGFR2 kinase inhibitors.
J.Med.Chem., 48, 2005
|
|
1Y6B
 
 | | Crystal structure of VEGFR2 in complex with a 2-anilino-5-aryl-oxazole inhibitor | | Descriptor: | N-(CYCLOPROPYLMETHYL)-4-(METHYLOXY)-3-({5-[3-(3-PYRIDINYL)PHENYL]-1,3-OXAZOL-2-YL}AMINO)BENZENESULFONAMIDE, Vascular endothelial growth factor receptor 2 | | Authors: | Harris, P.A, Cheung, M, Hunter, R.N, Brown, M.L, Veal, J.M, Nolte, R.T, Wang, L, Liu, W, Crosby, R.M, Johnson, J.H, Epperly, A.H, Kumar, R, Luttrell, D.K, Stafford, J.A. | | Deposit date: | 2004-12-05 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and evaluation of 2-anilino-5-aryloxazoles as a novel class of VEGFR2 kinase inhibitors.
J.Med.Chem., 48, 2005
|
|
1N7D
 
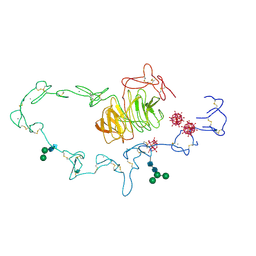 | | Extracellular domain of the LDL receptor | | Descriptor: | 12-TUNGSTOPHOSPHATE, CALCIUM ION, Low-density lipoprotein receptor, ... | | Authors: | Rudenko, G, Henry, L, Henderson, K, Ichtchenko, K, Brown, M.S, Goldstein, J.L, Deisenhofer, J. | | Deposit date: | 2002-11-13 | | Release date: | 2003-01-21 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of the LDL receptor extracellular domain at endosomal pH
Science, 298, 2002
|
|
1GNG
 
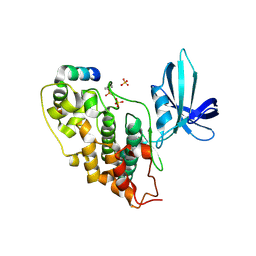 | | Glycogen synthase kinase-3 beta (GSK3) complex with FRATtide peptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FRATTIDE, GLYCOGEN SYNTHASE KINASE-3 BETA, ... | | Authors: | Bax, B, Carter, P.S, Lewis, C, Guy, A.R, Bridges, A, Tanner, R, Pettman, G, Mannix, C, Culbert, A.A, Brown, M.J.B, Smith, D.G, Reith, A.D. | | Deposit date: | 2001-10-04 | | Release date: | 2002-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structure of Phosphorylated Gsk-3Beta Complexed with a Peptide, Frattide, that Inhibits Beta-Catenin Phosphorylation
Structure, 9, 2001
|
|
5A2E
 
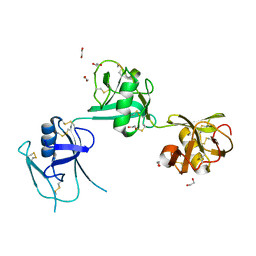 | | Extracellular SRCR domains of human CD6 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, T-CELL DIFFERENTIATION ANTIGEN CD6 | | Authors: | Chappell, P.E, Johnson, S, Lea, S.M, Brown, M.H. | | Deposit date: | 2015-05-18 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structures of Cd6 and its Ligand Cd166 Give Insight Into Their Interaction.
Structure, 23, 2015
|
|
1JT0
 
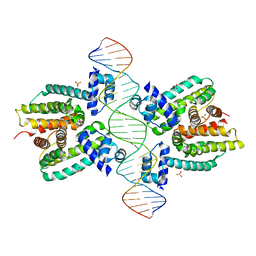 | | Crystal structure of a cooperative QacR-DNA complex | | Descriptor: | HYPOTHETICAL TRANSCRIPTIONAL REGULATOR IN QACA 5'REGION, QACA operator, SULFATE ION | | Authors: | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | Deposit date: | 2001-08-20 | | Release date: | 2002-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR.
EMBO J., 21, 2002
|
|
1JUP
 
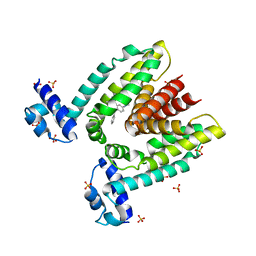 | | Crystal structure of the multidrug binding transcriptional repressor QacR bound to malachite green | | Descriptor: | HYPOTHETICAL TRANSCRIPTIONAL REGULATOR IN QACA 5'REGION, MALACHITE GREEN, SULFATE ION | | Authors: | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | Deposit date: | 2001-08-24 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural mechanisms of QacR induction and multidrug recognition.
Science, 294, 2001
|
|
