2GKJ
 
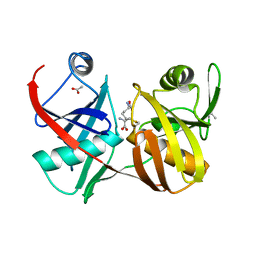 | | Crystal structure of diaminopimelate epimerase in complex with an irreversible inhibitor DL-AZIDAP | | Descriptor: | (2R,6S)-2,6-DIAMINO-2-METHYLHEPTANEDIOIC ACID, ACETIC ACID, Diaminopimelate epimerase | | Authors: | Pillai, B, Cherney, M.M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C, James, M.N. | | Deposit date: | 2006-04-02 | | Release date: | 2006-05-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into stereochemical inversion by diaminopimelate epimerase: An antibacterial drug target.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4M0D
 
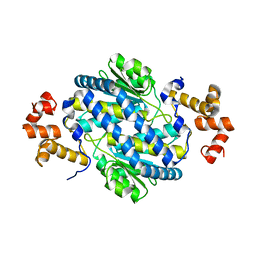 | |
2GKE
 
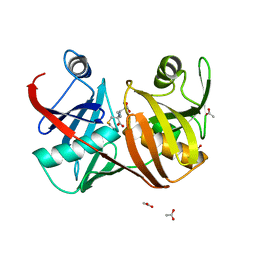 | | Crystal structure of diaminopimelate epimerase in complex with an irreversible inhibitor LL-AziDAP | | Descriptor: | (2S,6S)-2,6-DIAMINO-2-METHYLHEPTANEDIOIC ACID, ACETIC ACID, Diaminopimelate epimerase, ... | | Authors: | Pillai, B, Cherney, M.M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C, James, M.N. | | Deposit date: | 2006-04-01 | | Release date: | 2006-05-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into stereochemical inversion by diaminopimelate epimerase: An antibacterial drug target.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1KGQ
 
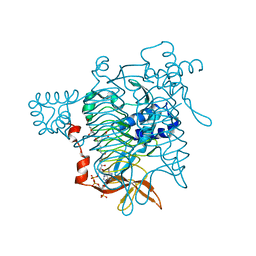 | | Crystal Structure of Tetrahydrodipicolinate N-Succinyltransferase in Complex with L-2-aminopimelate and Succinamide-CoA | | Descriptor: | (2S)-2-aminoheptanedioic acid, 2,3,4,5-TETRAHYDROPYRIDINE-2-CARBOXYLATE N-SUCCINYLTRANSFERASE, SUCCINAMIDE-COA | | Authors: | Beaman, T.W, Vogel, K.W, Drueckhammer, D.G, Blanchard, J.S, Roderick, S.L. | | Deposit date: | 2001-11-28 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Acyl group specificity at the active site of tetrahydridipicolinate N-succinyltransferase.
Protein Sci., 11, 2002
|
|
1KGT
 
 | | Crystal Structure of Tetrahydrodipicolinate N-Succinyltransferase in Complex with Pimelate and Succinyl-CoA | | Descriptor: | 2,3,4,5-TETRAHYDROPYRIDINE-2-CARBOXYLATE N-SUCCINYLTRANSFERASE, PIMELIC ACID, SUCCINYL-COENZYME A | | Authors: | Beaman, T.W, Vogel, K.W, Drueckhammer, D.G, Blanchard, J.S, Roderick, S.L. | | Deposit date: | 2001-11-28 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Acyl group specificity at the active site of tetrahydridipicolinate N-succinyltransferase.
Protein Sci., 11, 2002
|
|
1G2O
 
 | | CRYSTAL STRUCTURE OF PURINE NUCLEOSIDE PHOSPHORYLASE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH A TRANSITION-STATE INHIBITOR | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, PHOSPHATE ION, PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Shi, W, Basso, L.A, Tyler, P.C, Furneaux, R.H, Blanchard, J.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2000-10-20 | | Release date: | 2001-08-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of purine nucleoside phosphorylase from Mycobacterium tuberculosis in complexes with immucillin-H and its pieces.
Biochemistry, 40, 2001
|
|
1TDT
 
 | |
4JLF
 
 | | Inhibitor resistant (R220A) substitution in the Mycobacterium tuberculosis beta-lactamase | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Hazra, S, Kurz, S, Blanchard, J, Bonomo, R. | | Deposit date: | 2013-03-12 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Can inhibitor-resistant substitutions in the Mycobacterium tuberculosis beta-Lactamase BlaC lead to clavulanate resistance?: a biochemical rationale for the use of beta-lactam-beta-lactamase inhibitor combinations.
Antimicrob.Agents Chemother., 57, 2013
|
|
2BUE
 
 | | Structure of AAC(6')-Ib in complex with Ribostamycin and Coenzyme A. | | Descriptor: | AAC(6')-IB, CALCIUM ION, COENZYME A, ... | | Authors: | Vetting, M.W, Park, C.H, Hedge, S.S, Hooper, D.C, Blanchard, J.S. | | Deposit date: | 2008-03-20 | | Release date: | 2008-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanistic and Structural Analysis of Aminoglycoside N-Acetyltransferase Aac(6')-Ib and its Bifunctional, Fluoroquinolone-Active Aac(6')-Ib-Cr Variant.
Biochemistry, 47, 2008
|
|
2CIG
 
 | | Dihydrofolate reductase from Mycobacterium tuberculosis inhibited by the acyclic 4R isomer of INH-NADP a derivative of the prodrug isoniazid. | | Descriptor: | (4R)-ISONICOTINIC-ACETYL-NICOTINAMIDE-ADENINE DINUCLEOTIDE, DIHYDROFOLATE REDUCTASE, GLYCEROL, ... | | Authors: | Argyrou, A, Vetting, M.W, Aladegbami, B, Blanchard, J.S. | | Deposit date: | 2006-03-20 | | Release date: | 2006-04-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mycobacterium Tuberculosis Dihydrofolate Reductase is a Target for Isoniazid
Nat.Struct.Mol.Biol., 13, 2006
|
|
2BM5
 
 | | The Structure of MfpA (Rv3361c, P21 Crystal form). The Pentapeptide Repeat Protein from Mycobacterium tuberculosis Folds as A Right- handed Quadrilateral Beta-helix. | | Descriptor: | PENTAPEPTIDE REPEAT FAMILY PROTEIN, SULFATE ION | | Authors: | Hegde, S.S, Vetting, M.W, Roderick, S.L, Mitchenall, L.A, Maxwell, A, Takiff, H.E, Blanchard, J.S. | | Deposit date: | 2005-03-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Fluroquinolone Resistance Protein from Mycobacterium Tuberculosis that Mimics DNA
Science, 308, 2005
|
|
2BM6
 
 | | The Structure of MfpA (Rv3361c, C2221 Crystal form). The Pentapeptide Repeat Protein from Mycobacterium tuberculosis Folds as A Right- handed Quadrilateral Beta-helix. | | Descriptor: | CESIUM ION, PENTAPEPTIDE REPEAT FAMILY PROTEIN | | Authors: | Hegde, S.S, Vetting, M.W, Roderick, S.L, Mitchenall, L.A, Maxwell, A, Takiff, H.E, Blanchard, J.S. | | Deposit date: | 2005-03-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Fluroquinolone Resistance Protein from Mycobacterium Tuberculosis that Mimics DNA
Science, 308, 2005
|
|
1Q5N
 
 | |
2BM4
 
 | | The Structure of MfpA (Rv3361c, C2 Crystal form). The Pentapeptide Repeat Protein from Mycobacterium tuberculosis Folds as A Right- handed Quadrilateral Beta-helix. | | Descriptor: | PENTAPEPTIDE REPEAT FAMILY PROTEIN | | Authors: | Hegde, S.S, Vetting, M.W, Roderick, S.L, Mitchenall, L.A, Maxwell, A, Takiff, H.E, Blanchard, J.S. | | Deposit date: | 2005-03-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Fluroquinolone Resistance Protein from Mycobacterium Tuberculosis that Mimics DNA
Science, 308, 2005
|
|
2BM7
 
 | | The Structure of MfpA (Rv3361c, P3221 Crystal form). The Pentapeptide Repeat Protein from Mycobacterium tuberculosis Folds as A Right- handed Quadrilateral Beta-helix. | | Descriptor: | PENTAPEPTIDE REPEAT FAMILY PROTEIN | | Authors: | Hegde, S.S, Vetting, M.W, Roderick, S.L, Mitchenall, L.A, Maxwell, A, Takiff, H.E, Blanchard, J.S. | | Deposit date: | 2005-03-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Fluroquinolone Resistance Protein from Mycobacterium Tuberculosis that Mimics DNA
Science, 308, 2005
|
|
3NDE
 
 | |
3NCK
 
 | |
3NDG
 
 | |
3NBL
 
 | | Crystal Structure of BlaC-E166A covalently bound with Cefuroxime | | Descriptor: | (2R)-5-[(carbamoyloxy)methyl]-2-[(1R)-1-{[(2Z)-2-(furan-2-yl)-2-(methoxyimino)acetyl]amino}-2-oxoethyl]-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase | | Authors: | Tremblay, L.W, Blanchard, J.S. | | Deposit date: | 2010-06-03 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | BlaC-E166A covalently bound with cephalosporins and penicillins
To be Published
|
|
3N8S
 
 | | Crystal Structure of BlaC-E166A covalently bound with Cefamandole | | Descriptor: | (2R)-2-[(1R)-1-{[(2R)-2-hydroxy-2-phenylacetyl]amino}-2-oxoethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Tremblay, L.W, Blanchard, J.S. | | Deposit date: | 2010-05-28 | | Release date: | 2010-11-24 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Michaelis Complex (1.2 A) and the Covalent Acyl Intermediate (2.0 A) of Cefamandole Bound in the Active Sites of the Mycobacterium tuberculosis beta-Lactamase K73A and E166A Mutants.
Biochemistry, 49, 2010
|
|
3N6I
 
 | |
2AMA
 
 | | Crystal structure of human androgen receptor ligand binding domain in complex with dihydrotestosterone | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, SULFATE ION | | Authors: | Pereira de Jesus-Tran, K, Cote, P.-L, Cantin, L, Blanchet, J, Labrie, F, Breton, R. | | Deposit date: | 2005-08-09 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparison of crystal structures of human androgen receptor ligand-binding domain complexed with various agonists reveals molecular determinants responsible for binding affinity.
Protein Sci., 15, 2006
|
|
2AM9
 
 | | Crystal structure of human androgen receptor ligand binding domain in complex with testosterone | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Androgen receptor, GLYCEROL, ... | | Authors: | Pereira de Jesus-Tran, K, Cote, P.-L, Cantin, L, Blanchet, J, Labrie, F, Breton, R. | | Deposit date: | 2005-08-09 | | Release date: | 2006-05-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Comparison of crystal structures of human androgen receptor ligand-binding domain complexed with various agonists reveals molecular determinants responsible for binding affinity.
Protein Sci., 15, 2006
|
|
3M6B
 
 | | Crystal Structure of the Ertapenem Pre-isomerized Covalent Adduct with TB B-lactamase | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Tremblay, L.W, Fan, F, Blanchard, J.S. | | Deposit date: | 2010-03-15 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Biochemical and structural characterization of Mycobacterium tuberculosis beta-lactamase with the carbapenems ertapenem and doripenem.
Biochemistry, 49, 2010
|
|
3N8R
 
 | |
