6T8Q
 
 | | HKATII IN COMPLEX WITH LIGAND (2R)-N-benzyl-1-[6-methyl-5-(oxan-4-yl)-7-oxo-6H,7H-[1,3]thiazolo[5,4-d]pyrimidin-2-yl]pyrrolidine-2-carboxamide | | Descriptor: | (2~{R})-1-[6-methyl-5-(oxan-4-yl)-7-oxidanylidene-[1,3]thiazolo[5,4-d]pyrimidin-2-yl]-~{N}-(phenylmethyl)pyrrolidine-2-carboxamide, ACETATE ION, CADMIUM ION, ... | | Authors: | Blaesse, M, Venalainen, J. | | Deposit date: | 2019-10-24 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of sulfonamides and 9-oxo-2,8-diazaspiro[5,5]undecane-2-carboxamides as human kynurenine aminotransferase 2 (KAT2) inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
8VXE
 
 | | Structure of p38 alpha (Mitogen-activated protein kinase 14) complexed with inhibitor 6 | | Descriptor: | (4M)-4-[3-(4-fluorophenyl)-1-methyl-1H-pyrazol-4-yl]-1H-pyrrolo[2,3-b]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Blaesse, M, Steinbacher, S, Shaffer, P.L, Sharma, S, Thompson, A.A. | | Deposit date: | 2024-02-04 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Optimization of Selective and Brain Penetrant CK1 delta Inhibitors for the Treatment of Circadian Disruptions.
Acs Med.Chem.Lett., 15, 2024
|
|
1G5Q
 
 | | EPID H67N COMPLEXED WITH SUBSTRATE PEPTIDE DSYTC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, EPIDERMIN MODIFYING ENZYME EPID, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Blaesse, M, Kupke, T, Huber, R, Steinbacher, S. | | Deposit date: | 2000-11-02 | | Release date: | 2001-05-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal structure of the peptidyl-cysteine decarboxylase EpiD complexed with a pentapeptide substrate.
EMBO J., 19, 2000
|
|
6T8P
 
 | | HKATII IN COMPLEX WITH LIGAND (2R)-N-benzyl-1-[6-methyl-5-(oxan-4-yl)-7-oxo-6H,7H-[1,3]thiazolo[5,4-d]pyrimidin-2-yl]pyrrolidine-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 3,5-bis(fluoranyl)-~{N}-[5-[(2~{R})-2-(3-fluorophenyl)-3-methyl-butyl]-1,3,4-thiadiazol-2-yl]benzenesulfonamide, IODIDE ION, ... | | Authors: | Blaesse, M, Venalainen, J. | | Deposit date: | 2019-10-24 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Discovery of sulfonamides and 9-oxo-2,8-diazaspiro[5,5]undecane-2-carboxamides as human kynurenine aminotransferase 2 (KAT2) inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
1G63
 
 | | PEPTIDYL-CYSTEINE DECARBOXYLASE EPID | | Descriptor: | EPIDERMIN MODIFYING ENZYME EPID, FLAVIN MONONUCLEOTIDE | | Authors: | Blaesse, M, Kupke, T, Huber, R, Steinbac, S. | | Deposit date: | 2000-11-03 | | Release date: | 2001-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the peptidyl-cysteine decarboxylase EpiD complexed with a pentapeptide substrate.
EMBO J., 19, 2000
|
|
1P3Y
 
 | | MrsD from Bacillus sp. HIL-Y85/54728 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MrsD protein | | Authors: | Blaesse, M, Kupke, T, Huber, R, Steinbacher, S. | | Deposit date: | 2003-04-19 | | Release date: | 2003-08-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structure of MrsD, an FAD-binding protein of the HFCD family.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
6E5B
 
 | | Human Immunoproteasome 20S particle in complex with compound 1 | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Steinbacher, S, Augustin, M, Blaesse, M, Harris, S.F. | | Deposit date: | 2018-07-19 | | Release date: | 2019-09-11 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Design and Evaluation of Highly Selective Human Immunoproteasome Inhibitors Reveal a Compensatory Process That Preserves Immune Cell Viability.
J.Med.Chem., 62, 2019
|
|
6B8A
 
 | | Crystal structure of MvfR ligand binding domain in complex with M64 | | Descriptor: | 2-[(5-nitro-1H-benzimidazol-2-yl)sulfanyl]-N-(4-phenoxyphenyl)acetamide, COBALT HEXAMMINE(III), DNA-binding transcriptional regulator | | Authors: | Kitao, T, Steinbacher, S, Maskos, K, Blaesse, M, Rahme, L.G. | | Deposit date: | 2017-10-05 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Molecular Insights into Function and Competitive Inhibition ofPseudomonas aeruginosaMultiple Virulence Factor Regulator.
MBio, 9, 2018
|
|
1ZRZ
 
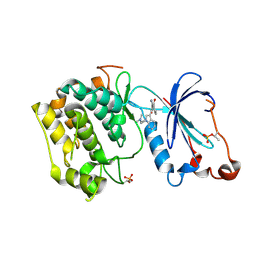 | | Crystal Structure of the Catalytic Domain of Atypical Protein Kinase C-iota | | Descriptor: | 3-{1-[3-(DIMETHYLAMINO)PROPYL]-1H-INDOL-3-YL}-4-(1H-INDOL-3-YL)-1H-PYRROLE-2,5-DIONE, Protein kinase C, iota | | Authors: | Messerschmidt, A, Macieira, S, Velarde, M, Baedeker, M, Benda, C, Jestel, A, Brandstetter, H, Neuefeind, T, Blaesse, M, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-05-23 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Atypical Protein Kinase C-iota Reveals Interaction Mode of Phosphorylation Site in Turn Motif
J.Mol.Biol., 352, 2005
|
|
3RGF
 
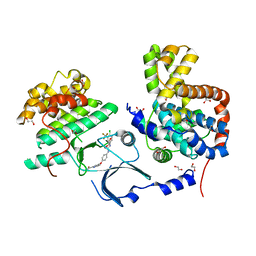 | | Crystal Structure of human CDK8/CycC | | Descriptor: | 1,2-ETHANEDIOL, 4-{4-[({[4-CHLORO-3-(TRIFLUOROMETHYL)PHENYL]AMINO}CARBONYL)AMINO]PHENOXY}-N-METHYLPYRIDINE-2-CARBOXAMIDE, Cyclin-C, ... | | Authors: | Schneider, E.V, Boettcher, J, Blaesse, M, Huber, R, Maskos, K. | | Deposit date: | 2011-04-08 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of CDK8/CycC Implicates Specificity in the CDK/Cyclin Family and Reveals Interaction with a Deep Pocket Binder.
J.Mol.Biol., 412, 2011
|
|
1MVN
 
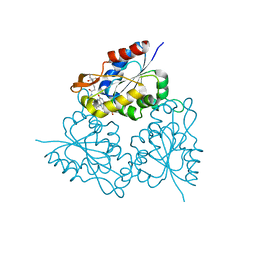 | | PPC decarboxylase mutant C175S complexed with pantothenoylaminoethenethiol | | Descriptor: | 2,4-DIHYDROXY-N-[2-(2-MERCAPTO-VINYLCARBAMOYL)-ETHYL]-3,3-DIMETHYL-BUTYRAMIDE, FLAVIN MONONUCLEOTIDE, PPC decarboxylase AtHAL3a | | Authors: | Steinbacher, S, Hernandez-Acosta, P, Bieseler, B, Blaesse, M, Huber, R, Culianez-Macia, F.A, Kupke, T. | | Deposit date: | 2002-09-26 | | Release date: | 2003-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal Structure of the Plant PPC Decarboxylase AtHAL3a Complexed with an Ene-thiol Reaction Intermediate
J.Mol.Biol., 327, 2003
|
|
1MVL
 
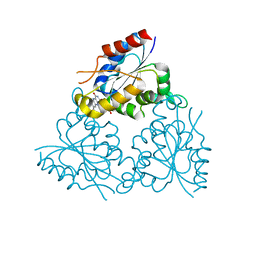 | | PPC decarboxylase mutant C175S | | Descriptor: | FLAVIN MONONUCLEOTIDE, PPC decarboxylase AtHAL3a | | Authors: | Steinbacher, S, Hernandez-Acosta, P, Bieseler, B, Blaesse, M, Huber, R, Culianez-Macia, F.A, Kupke, T. | | Deposit date: | 2002-09-26 | | Release date: | 2003-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Plant PPC Decarboxylase AtHAL3a Complexed with an Ene-thiol Reaction Intermediate
J.Mol.Biol., 327, 2003
|
|
4EAW
 
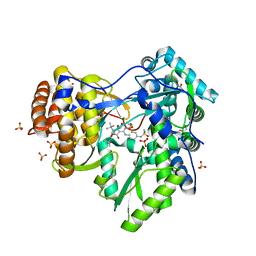 | | HCV NS5B in complex with IDX375 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, N-{(1S)-3-[(5S)-5-tert-butyl-1-(3,3-dimethylbutyl)-4-hydroxy-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]-1-ethoxy-1-oxido-1,4-dihydro-2,4,1-benzodiazaphosphinin-7-yl}methanesulfonamide, ... | | Authors: | Dousson, C.B, Paparin, J.-L, Surleraux, D, Augustin, M, Blaesse, M, Hoeppner, S, Krapp, S, Wenzkowski, C. | | Deposit date: | 2012-03-22 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HCV NS5B in complex with IDX375
To be Published
|
|
5WR7
 
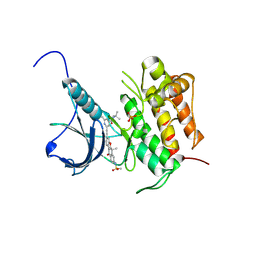 | | Crystal structure of Trk-A complexed with a selective inhibitor CH7057288 | | Descriptor: | High affinity nerve growth factor receptor, N-tert-butyl-2-[2-[6,6-dimethyl-8-(methylsulfonylamino)-11-oxidanylidene-naphtho[2,3-b][1]benzofuran-3-yl]ethynyl]-6-methyl-pyridine-4-carboxamide | | Authors: | Tanaka, H, Blaesse, M, Augustin, M, Goesser, C. | | Deposit date: | 2016-11-30 | | Release date: | 2017-12-06 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Selective TRK Inhibitor CH7057288 against TRK Fusion-Driven Cancer.
Mol. Cancer Ther., 17, 2018
|
|
5M6U
 
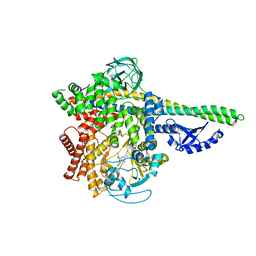 | | HUMAN PI3KDELTA IN COMPLEX WITH LASW1579 | | Descriptor: | 4-azanyl-6-[[(1~{S})-1-(4-oxidanylidene-3-phenyl-pyrrolo[2,1-f][1,2,4]triazin-2-yl)ethyl]amino]pyrimidine-5-carbonitrile, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Segarra, V, Hernandez, B, Lozoya, E, Blaesse, M, Hoeppner, S, Jestel, A. | | Deposit date: | 2016-10-26 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of a Potent, Selective, and Orally Available PI3K delta Inhibitor for the Treatment of Inflammatory Diseases.
ACS Med Chem Lett, 8, 2017
|
|
4F4P
 
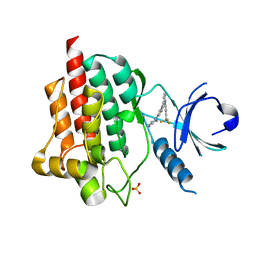 | | SYK in COMPLEX WITH LIGAND LASW836 | | Descriptor: | N-{6-[3-(piperazin-1-yl)phenyl]pyridin-2-yl}-4-(trifluoromethyl)pyridin-2-amine, SULFATE ION, Tyrosine-protein kinase SYK | | Authors: | Lopez, M, Segarra, V, Vidal, B, Wenzkowski, C, Jestel, A, Krapp, S, Blaesse, M, Nagel, S, Schreiner, P. | | Deposit date: | 2012-05-11 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Highly potent aminopyridines as Syk kinase inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1G8F
 
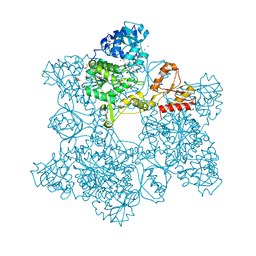 | | ATP SULFURYLASE FROM S. CEREVISIAE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, CADMIUM ION, ... | | Authors: | Ullrich, T.C, Blaesse, M, Huber, R. | | Deposit date: | 2000-11-17 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of ATP sulfurylase from Saccharomyces cerevisiae, a key enzyme in sulfate activation.
EMBO J., 20, 2001
|
|
1G8G
 
 | | ATP SULFURYLASE FROM S. CEREVISIAE: THE BINARY PRODUCT COMPLEX WITH APS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ADENOSINE-5'-PHOSPHOSULFATE, ... | | Authors: | Ullrich, T.C, Blaesse, M, Huber, R. | | Deposit date: | 2000-11-17 | | Release date: | 2001-05-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of ATP sulfurylase from Saccharomyces cerevisiae, a key enzyme in sulfate activation.
EMBO J., 20, 2001
|
|
1G8H
 
 | | ATP SULFURYLASE FROM S. CEREVISIAE: THE TERNARY PRODUCT COMPLEX WITH APS AND PPI | | Descriptor: | ACETIC ACID, ADENOSINE-5'-PHOSPHOSULFATE, CADMIUM ION, ... | | Authors: | Ullrich, T.C, Blaesse, M, Huber, R. | | Deposit date: | 2000-11-17 | | Release date: | 2001-05-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ATP sulfurylase from Saccharomyces cerevisiae, a key enzyme in sulfate activation.
EMBO J., 20, 2001
|
|
6HGY
 
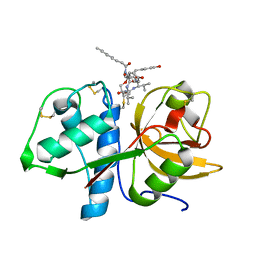 | | CRYSTAL STRUCTURE OF CATHEPSIN K WITH N-DESMETHYL THALASSOSPIRAMIDE C | | Descriptor: | Cathepsin K, THALASSOSPIRAMIDE C | | Authors: | Zakarian, A, Buckman, B.O, Adler, M, Griessner, A, Blaesse, M. | | Deposit date: | 2018-08-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Total Synthesis of Covalent Cysteine Protease Inhibitor N-Desmethyl Thalassospiramide C and Crystallographic Evidence for Its Mode of Action.
Org.Lett., 21, 2019
|
|
6Y9S
 
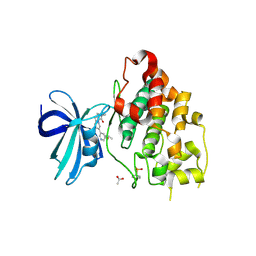 | | Crystal structure of GSK-3b in complex with the imidazo[1,5-a]pyridine-3-carboxamide inhibitor 16 | | Descriptor: | ACETATE ION, Glycogen synthase kinase-3 beta, ~{N}-(oxan-4-ylmethyl)-6-(5-propan-2-yloxypyridin-3-yl)imidazo[1,5-a]pyridine-3-carboxamide | | Authors: | Krapp, S, Griessner, A, Blaesse, M, Buonfiglio, R, Ombrato, R. | | Deposit date: | 2020-03-10 | | Release date: | 2020-05-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery of Novel Imidazopyridine GSK-3 beta Inhibitors Supported by Computational Approaches.
Molecules, 25, 2020
|
|
6Y9R
 
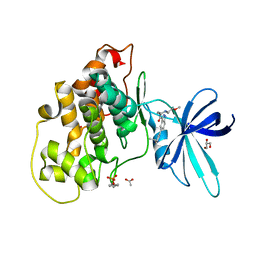 | | Crystal structure of GSK-3b in complex with the 1H-indazole-3-carboxamide inhibitor 2 | | Descriptor: | ACETATE ION, GLYCEROL, Glycogen synthase kinase-3 beta, ... | | Authors: | Krapp, S, Griessner, A, Blaesse, M, Buonfiglio, R, Ombrato, R. | | Deposit date: | 2020-03-10 | | Release date: | 2020-05-20 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of Novel Imidazopyridine GSK-3 beta Inhibitors Supported by Computational Approaches.
Molecules, 25, 2020
|
|
5LLS
 
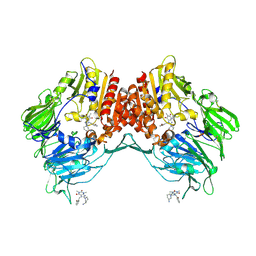 | | Porcine dipeptidyl peptidase IV in complex with 8-(3-aminopiperidin-1-yl)-7-[(2-bromophenyl)methyl]-1,3-dimethyl-2,3,6,7-tetrahydro-1H-purine-2,6-dione | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 8-[(3~{R})-3-azanylpiperidin-1-yl]-7-[(2-bromophenyl)methyl]-1,3-dimethyl-purine-2,6-dione, ... | | Authors: | Nar, H, Blaesse, M. | | Deposit date: | 2016-07-28 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Comparative Analysis of Binding Kinetics and Thermodynamics of Dipeptidyl Peptidase-4 Inhibitors and Their Relationship to Structure.
J.Med.Chem., 59, 2016
|
|
7L5O
 
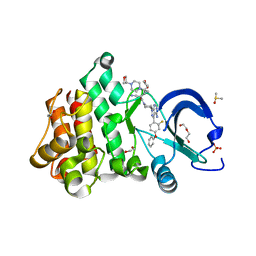 | | Crystal structure of the noncovalently bonded complex of rilzabrutinib with BTK | | Descriptor: | (2E)-2-{(3R)-3-[4-amino-3-(2-fluoro-4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidine-1-carbonyl}-4-methyl-4-[4-(oxetan-3-yl)piperazin-1-yl]pent-2-enenitrile, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Bradshaw, J.M, Brameld, K.A, Mrosek, M, Lammens, A, Blaesse, M. | | Deposit date: | 2020-12-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | The Discovery Rilzabrutinib (PRN1008): A Reversible Covalent BTK Inhibitor for Immune Mediated Diseases
To Be Published
|
|
7L5P
 
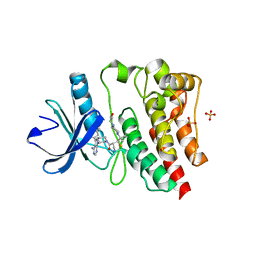 | | Crystal structure of the covalently bonded complex of rilzabrutinib with BTK | | Descriptor: | (2E)-2-{(3R)-3-[4-amino-3-(2-fluoro-4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidine-1-carbonyl}-4-methyl-4-[4-(oxetan-3-yl)piperazin-1-yl]pent-2-enenitrile, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Bradshaw, J.M, Brameld, K.A, Mrosek, M, Lammens, A, Blaesse, M. | | Deposit date: | 2020-12-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The Discovery Rilzabrutinib (PRN1008): A Reversible Covalent BTK Inhibitor for Immune Mediated Diseases
To Be Published
|
|
