5IBS
 
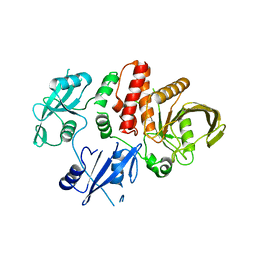 | | Structure of E76Q, a Cancer-Associated Mutation of the Oncogenic Phosphatase SHP2 | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Blacklow, S.C, Stams, T, Fodor, M, LaRochelle, J.R. | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural and Functional Consequences of Three Cancer-Associated Mutations of the Oncogenic Phosphatase SHP2.
Biochemistry, 55, 2016
|
|
5IBM
 
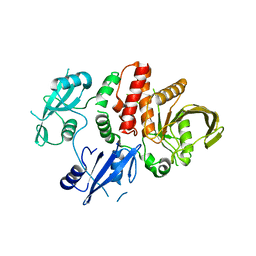 | | Structure of S502P, a Cancer-Associated Mutation of the Oncogenic Phosphatase SHP2 | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Blacklow, S.C, Stams, T, Fodor, M, LaRochelle, J.R. | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural and Functional Consequences of Three Cancer-Associated Mutations of the Oncogenic Phosphatase SHP2.
Biochemistry, 55, 2016
|
|
6BN5
 
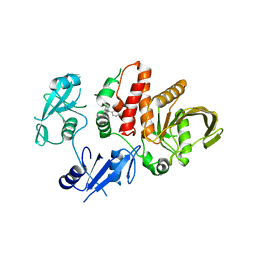 | | Non-receptor Protein Tyrosine Phosphatase SHP2 F285S in Complex with Allosteric Inhibitor JLR-2 | | Descriptor: | 3-benzyl-8-chloro-2-hydroxy-4H-pyrimido[2,1-b][1,3]benzothiazol-4-one, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Blacklow, S.C, Stams, T, Fodor, M, LaRochelle, J.R. | | Deposit date: | 2017-11-16 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Identification of an allosteric benzothiazolopyrimidone inhibitor of the oncogenic protein tyrosine phosphatase SHP2.
Bioorg. Med. Chem., 25, 2017
|
|
8ESV
 
 | |
5TCX
 
 | | Crystal structure of human tetraspanin CD81 | | Descriptor: | CD81 antigen, CHOLESTEROL | | Authors: | Zimmerman, B, McMillan, B.J, Seegar, T.C.M, Kruse, A.C, Blacklow, S.C. | | Deposit date: | 2016-09-16 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | Crystal Structure of a Full-Length Human Tetraspanin Reveals a Cholesterol-Binding Pocket.
Cell, 167, 2016
|
|
3I08
 
 | |
3NBN
 
 | |
8V0E
 
 | | ANK repeat of MIB1 | | Descriptor: | E3 ubiquitin-protein ligase MIB1, Unidentified MIB1 peptide | | Authors: | Cao, R, Blacklow, S.C. | | Deposit date: | 2023-11-17 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural requirements for activity of Mind bomb1 in Notch signaling.
Structure, 32, 2024
|
|
8V0D
 
 | | Ubch5B-RING3 of MIB1 fusion structure | | Descriptor: | Ubch5B-RING3 of MIB1 fusion protein, ZINC ION | | Authors: | Cao, R, Blacklow, S.C. | | Deposit date: | 2023-11-17 | | Release date: | 2024-11-27 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural requirements for activity of Mind bomb1 in Notch signaling.
Structure, 32, 2024
|
|
8UO5
 
 | | Protein Phosphatase 2A B55 subunit in complex with IER5 | | Descriptor: | FE (III) ION, Immediate early response gene 5 protein, Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform, ... | | Authors: | Jones, D.T.D, Cao, R, Rawson, S.D, Blacklow, S.C. | | Deposit date: | 2023-10-19 | | Release date: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Protein Phosphatase 2A B55 subunit in complex with IER5
To Be Published
|
|
6PY8
 
 | | Crystal structure of the RBPJ-NOTCH1-NRARP ternary complex bound to DNA | | Descriptor: | DNA, Neurogenic locus notch homolog protein 1, Notch-regulated ankyrin repeat-containing protein, ... | | Authors: | Jarrett, S.M, Seegar, T.C.M, Blacklow, S.C. | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Extension of the Notch intracellular domain ankyrin repeat stack by NRARP promotes feedback inhibition of Notch signaling.
Sci.Signal., 12, 2019
|
|
7JIC
 
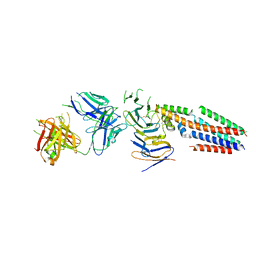 | | Structure of human CD19-CD81 co-receptor complex bound to coltuximab Fab fragment | | Descriptor: | B-lymphocyte antigen CD19, CD81 antigen, Coltuximab Heavy Chain, ... | | Authors: | Susa, K.J, Rawson, S, Kruse, A.C, Blacklow, S.C. | | Deposit date: | 2020-07-23 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the B cell co-receptor CD19 bound to the tetraspanin CD81
Science, 371, 2021
|
|
4XI7
 
 | |
4XIB
 
 | |
2FCW
 
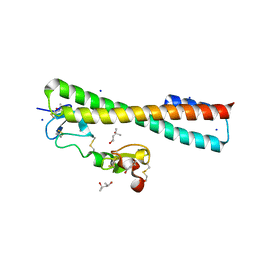 | | Structure of a Complex Between the Pair of the LDL Receptor Ligand-Binding Modules 3-4 and the Receptor Associated Protein (RAP). | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha-2-macroglobulin receptor-associated protein, CALCIUM ION, ... | | Authors: | Beglova, N, Fisher, C, Blacklow, S.C. | | Deposit date: | 2005-12-12 | | Release date: | 2006-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structure of an LDLR-RAP Complex Reveals a General Mode for Ligand Recognition by Lipoprotein Receptors
Mol.Cell, 22, 2006
|
|
2F8Y
 
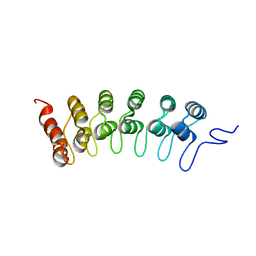 | | Crystal structure of human Notch1 ankyrin repeats to 1.55A resolution. | | Descriptor: | Notch homolog 1, translocation-associated (Drosophila), SULFATE ION | | Authors: | Nam, Y, Sliz, P, Blacklow, S.C. | | Deposit date: | 2005-12-04 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes.
Cell(Cambridge,Mass.), 124, 2006
|
|
4XI6
 
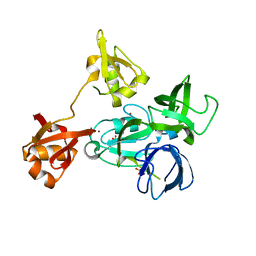 | |
2F8X
 
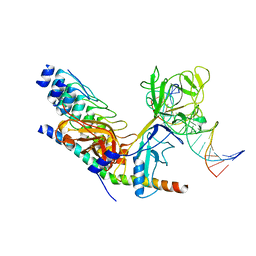 | | Crystal structure of activated Notch, CSL and MAML on HES-1 promoter DNA sequence | | Descriptor: | 5'-D(*GP*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*AP*AP*A)-3', 5'-D(*TP*TP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*TP*AP*AP*C)-3', Mastermind-like protein 1, ... | | Authors: | Nam, Y, Sliz, P, Blacklow, S.C. | | Deposit date: | 2005-12-04 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes.
Cell(Cambridge,Mass.), 124, 2006
|
|
5L2Q
 
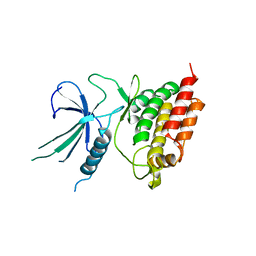 | |
6NCE
 
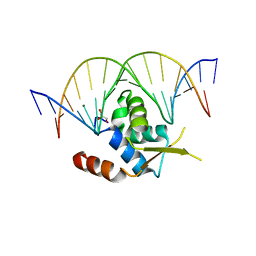 | | Crystal structure of the human FOXN3 DNA binding domain in complex with a forkhead DNA sequence | | Descriptor: | DNA (5'-D(*AP*CP*AP*TP*TP*GP*TP*TP*TP*AP*CP*TP*TP*AP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*TP*AP*AP*GP*TP*AP*AP*AP*CP*AP*AP*TP*G)-3'), Forkhead box protein N3, ... | | Authors: | Rogers, J.M, Jarrett, S.M, Seegar, T.C, Waters, C.T, Hallworth, A.N, Blacklow, S.C, Bulyk, M.L. | | Deposit date: | 2018-12-11 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Bispecific Forkhead Transcription Factor FoxN3 Recognizes Two Distinct Motifs with Different DNA Shapes.
Mol. Cell, 74, 2019
|
|
6NCM
 
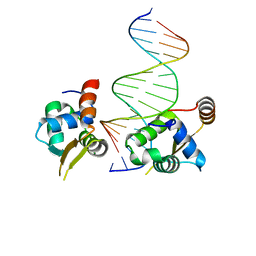 | | Crystal structure of the human FOXN3 DNA binding domain in complex with a forkhead-like (FHL) DNA sequence | | Descriptor: | DNA (5'-D(*AP*TP*AP*GP*CP*GP*TP*CP*TP*TP*AP*GP*CP*AP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*TP*GP*CP*TP*AP*AP*GP*AP*CP*GP*CP*TP*A)-3'), Forkhead box protein N3, ... | | Authors: | Rogers, J.M, Jarrett, S.M, Seegar, T.C, Waters, C.T, Hallworth, A.N, Blacklow, S.C, Bulyk, M.L. | | Deposit date: | 2018-12-11 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Bispecific Forkhead Transcription Factor FoxN3 Recognizes Two Distinct Motifs with Different DNA Shapes.
Mol. Cell, 74, 2019
|
|
7RDB
 
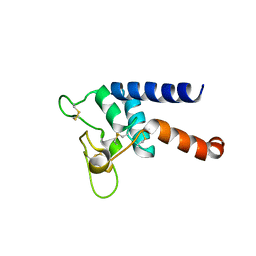 | | Crystal structure of Tspan15 large extracellular loop (Tspan15 LEL) | | Descriptor: | Tetraspanin-15 | | Authors: | Lipper, C.H, Gabriel, K.H, Seegar, T.C.M, Durr, K.L, Tomlinson, M.G, Blacklow, S.C. | | Deposit date: | 2021-07-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of the Tspan15 LEL domain reveals a conserved ADAM10 binding site.
Structure, 30, 2022
|
|
7RD5
 
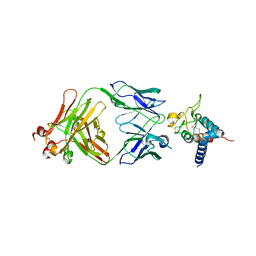 | | Crystal structure of Tspan15 large extracellular loop (Tspan15 LEL) in complex with 1C12 Fab | | Descriptor: | 1C12 Fab Heavy Chain, 1C12 Fab Light Chain, Tetraspanin-15 | | Authors: | Lipper, C.H, Gabriel, K.H, Seegar, T.C.M, Durr, K.L, Tomlinson, M.G, Blacklow, S.C. | | Deposit date: | 2021-07-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of the Tspan15 LEL domain reveals a conserved ADAM10 binding site.
Structure, 30, 2022
|
|
5HQG
 
 | |
5IGO
 
 | |
