2LUZ
 
 | | Solution NMR Structure of CalU16 from Micromonospora echinospora, Northeast Structural Genomics Consortium (NESG) Target MiR12 | | Descriptor: | CalU16 | | Authors: | Ramelot, T.A, Yang, Y, Lee, H, Pederson, K, Lee, D, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Wrobel, R.L, Bingman, C.A, Singh, S, Thorson, J.S, Prestegard, J.H, Montelione, G.T, Phillips Jr, G.N, Kennedy, M.A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-22 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure-Guided Functional Characterization of Enediyne Self-Sacrifice Resistance Proteins, CalU16 and CalU19.
Acs Chem.Biol., 9, 2014
|
|
4L7I
 
 | | Crystal structure of S-Adenosylmethionine synthase from Sulfolobus solfataricus complexed with SAM and PPi | | Descriptor: | DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-06-13 | | Release date: | 2013-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
4KX3
 
 | |
2O55
 
 | | Crystal Structure of a putative glycerophosphodiester phosphodiesterase from Galdieria sulphuraria | | Descriptor: | SULFATE ION, putative glycerophosphodiester phosphodiesterase | | Authors: | Mccoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-12-05 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Crystal Structure of a putative glycerophosphodiester phosphodiesterase from Galdieria sulphuraria
To be Published
|
|
2NYI
 
 | | Crystal Structure of an Unknown Protein from Galdieria sulphuraria | | Descriptor: | unknown protein | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, McCoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-11-20 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of tandem ACT domain-containing protein ACTP from Galdieria sulphuraria.
Proteins, 80, 2012
|
|
2NXF
 
 | | Crystal Structure of a dimetal phosphatase from Danio rerio LOC 393393 | | Descriptor: | ETHANOL, PHOSPHATE ION, Putative dimetal phosphatase, ... | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, McCoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-11-17 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a dimetal phosphatase from Danio rerio LOC 393393
To be Published
|
|
2O57
 
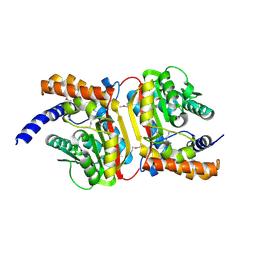 | | Crystal Structure of a putative sarcosine dimethylglycine methyltransferase from Galdieria sulphuraria | | Descriptor: | putative sarcosine dimethylglycine methyltransferase | | Authors: | Mccoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-12-05 | | Release date: | 2006-12-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.946 Å) | | Cite: | Crystal Structure of a putative sarcosine dimethylglycine methyltransferase from Galdieria sulphuraria
To be Published
|
|
2OA1
 
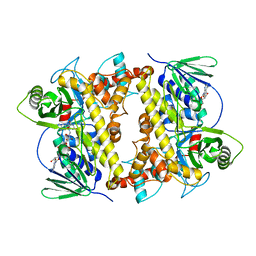 | | Crystal Structure of RebH, a FAD-dependent halogenase from Lechevalieria aerocolonigenes, the L-Tryptophan with FAD complex | | Descriptor: | ADENOSINE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bitto, E, Bingman, C.A, Singh, S, Phillips Jr, G.N. | | Deposit date: | 2006-12-14 | | Release date: | 2007-04-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of flavin-dependent tryptophan 7-halogenase RebH.
Proteins, 70, 2008
|
|
4KX5
 
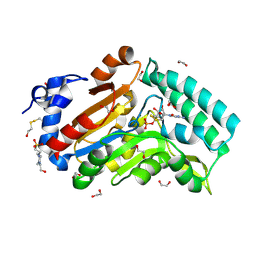 | |
4L2Z
 
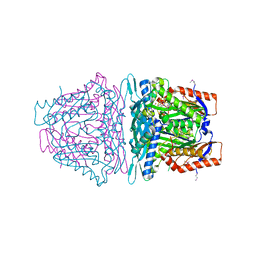 | | Crystal structure of S-Adenosylmethionine synthetase from Sulfolobus solfataricus complexed with SAE and PPi | | Descriptor: | DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-06-05 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
2O9Z
 
 | | Crystal Structure of RebH, a FAD-dependent halogenase from Lechevalieria aerocolonigenes, the Apo form | | Descriptor: | PHOSPHATE ION, Tryptophan halogenase | | Authors: | Bitto, E, Bingman, C.A, Phillips Jr, G.N. | | Deposit date: | 2006-12-14 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | The structure of flavin-dependent tryptophan 7-halogenase RebH.
Proteins, 70, 2008
|
|
4M83
 
 | | Ensemble refinement of protein crystal structure (2IYF) of macrolide glycosyltransferases OleD complexed with UDP and Erythromycin A | | Descriptor: | ERYTHROMYCIN A, MAGNESIUM ION, Oleandomycin glycosyltransferase, ... | | Authors: | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-08-12 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
4M60
 
 | | Crystal structure of macrolide glycosyltransferases OleD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Oleandomycin glycosyltransferase, SODIUM ION | | Authors: | Olmos Jr, J.L, Martinez III, E, Wang, F, Helmich, K.E, Singh, S, Xu, W, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-08-08 | | Release date: | 2013-09-04 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
4M7P
 
 | | Ensemble refinement of protein crystal structure of macrolide glycosyltransferases OleD | | Descriptor: | Oleandomycin glycosyltransferase, SODIUM ION | | Authors: | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-08-12 | | Release date: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
4WKY
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmN KS2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-ketoacyl synthase, GLYCEROL, ... | | Authors: | Cuff, M.E, Mack, J.C, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-10-03 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4FZR
 
 | | Crystal Structure of SsfS6, Streptomyces sp. SF2575 glycosyltransferase | | Descriptor: | SsfS6 | | Authors: | Wang, F, Zhou, M, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-07-07 | | Release date: | 2012-07-25 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Crystal structure of SsfS6, the putative C-glycosyltransferase involved in SF2575 biosynthesis.
Proteins, 81, 2013
|
|
4G2T
 
 | | Crystal Structure of Streptomyces sp. SF2575 glycosyltransferase SsfS6, complexed with thymidine diphosphate | | Descriptor: | SsfS6, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Wang, F, Zhou, M, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-07-12 | | Release date: | 2012-07-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Crystal structure of SsfS6, the putative C-glycosyltransferase involved in SF2575 biosynthesis.
Proteins, 81, 2013
|
|
4HPV
 
 | | Crystal structure of S-Adenosylmethionine synthetase from Sulfolobus solfataricus | | Descriptor: | S-adenosylmethionine synthase | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-10-24 | | Release date: | 2012-11-14 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.214 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
4FE3
 
 | |
4I4K
 
 | | Streptomyces globisporus C-1027 9-membered enediyne conserved protein SgcE6 | | Descriptor: | CITRIC ACID, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Kim, Y, Bigelow, L, Clancy, S, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-27 | | Release date: | 2012-12-12 | | Last modified: | 2016-12-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of SgcJ, an NTF2-like superfamily protein involved in biosynthesis of the nine-membered enediyne antitumor antibiotic C-1027.
J Antibiot (Tokyo), 69, 2016
|
|
2H39
 
 | | Crystal Structure of an ADP-Glucose Phosphorylase from Arabidopsis thaliana with bound ADP-Glucose | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE-GLUCOSE, CHLORIDE ION, Probable galactose-1-phosphate uridyl transferase, ... | | Authors: | McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-05-22 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal Structure of an ADP-Glucose Phosphorylase from Arabidopsis thaliana with bound ADP-Glucose
To be Published
|
|
4HX6
 
 | | Streptomyces globisporus C-1027 NADH:FAD oxidoreductase SgcE6 | | Descriptor: | ACETATE ION, Oxidoreductase, SULFATE ION | | Authors: | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-09 | | Release date: | 2012-11-28 | | Last modified: | 2016-12-07 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
4HVM
 
 | | Crystal structure of tallysomycin biosynthesis protein TlmII | | Descriptor: | SULFATE ION, TlmII | | Authors: | Chang, C, Bigelow, L, Bearden, J, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-06 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Crystal structure of tallysomycin biosynthesis protein TlmII
To be Published
|
|
4HZP
 
 | | The Structure of the Bifunctional Acetyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double Hot Dog Fold | | Descriptor: | Bifunctional methylmalonyl-CoA:ACP Acyltransferase/Decarboxylase, CHLORIDE ION, COENZYME A, ... | | Authors: | Lohman, J.R, Bingman, C.A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-15 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of the Bifunctional Acyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double-Hot-Dog Fold.
Biochemistry, 52, 2013
|
|
4HZN
 
 | | The Structure of the Bifunctional Acetyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double Hot Dog Fold | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional Methylmalonyl-CoA:ACP Acyltransferase/Decarboxylase, GLYCEROL, ... | | Authors: | Lohman, J.R, Bingman, C.A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-15 | | Release date: | 2013-01-30 | | Last modified: | 2013-02-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the Bifunctional Acyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double-Hot-Dog Fold.
Biochemistry, 52, 2013
|
|
