1Q7G
 
 | | Homoserine Dehydrogenase in complex with suicide inhibitor complex NAD-5-hydroxy-4-Oxonorvaline | | Descriptor: | Homoserine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE-5-HYDROXY-4-OXONORVALINE, SODIUM ION | | Authors: | Jacques, S.L, Mirza, I.A, Ejim, L, Koteva, K, Hughes, D.W, Green, K, Kinach, R, Honek, J.F, Lai, H.K, Berghuis, A.M, Wright, G.D. | | Deposit date: | 2003-08-18 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Enzyme assisted suicide: Molecular basis for the antifungal activity of 5-hydroxy-4-oxonorvaline by potent inhibition of homoserine dehydrogenase
Chem.Biol., 10, 2003
|
|
4XJE
 
 | |
4XQR
 
 | |
4XQT
 
 | |
4XQS
 
 | |
2B0Q
 
 | |
2B7L
 
 | | Crystal Structure of CTP:Glycerol-3-Phosphate Cytidylyltransferase from Staphylococcus aureus | | Descriptor: | glycerol-3-phosphate cytidylyltransferase | | Authors: | Fong, D.H, Yim, V.C.-N, D'Elia, M.A, Brown, E.D, Berghuis, A.M. | | Deposit date: | 2005-10-04 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of CTP:glycerol-3-phosphate cytidylyltransferase from Staphylococcus aureus: examination of structural basis for kinetic mechanism.
Biochim.Biophys.Acta, 1764, 2006
|
|
2B61
 
 | | Crystal Structure of Homoserine Transacetylase | | Descriptor: | Homoserine O-acetyltransferase | | Authors: | Mirza, I.A, Nazi, I, Korczynska, M, Wright, G.D, Berghuis, A.M. | | Deposit date: | 2005-09-29 | | Release date: | 2005-11-15 | | Last modified: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Homoserine Transacetylase from Haemophilus influenzae Reveals a New Family of alpha/beta-Hydrolases
Biochemistry, 44, 2005
|
|
1B87
 
 | |
2Z2O
 
 | |
2Z2N
 
 | |
2Z2P
 
 | | Crystal Structure of catalytically inactive H270A virginiamycin B lyase from Staphylococcus aureus with Quinupristin | | Descriptor: | 5-(2-DIETHYLAMINO-ETHANESULFONYL)-21-HYDROXY-10-ISOPROPYL-11,19-DIMETHYL-9,26-DIOXA-3,15,28-TRIAZA-TRICYCLO[23.2.1.00,255]OCTACOSA-1(27),12,17,19,25(28)-PENTAENE-2,8,14,23-TETRAONE, MAGNESIUM ION, Quinupristin, ... | | Authors: | Korczynska, M, Berghuis, A.M. | | Deposit date: | 2007-05-25 | | Release date: | 2007-06-19 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Streptogramin B Resistance in Staphylococcus Aureus by Virginiamycin B Lyase
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1SB9
 
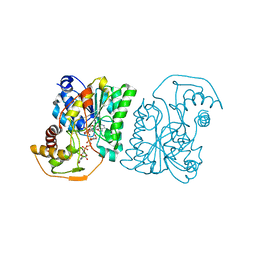 | | Crystal structure of Pseudomonas aeruginosa UDP-N-acetylglucosamine 4-epimerase complexed with UDP-glucose | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-5'-DIPHOSPHATE-GLUCOSE, wbpP | | Authors: | Ishiyama, N, Creuzenet, C, Lam, J.S, Berghuis, A.M. | | Deposit date: | 2004-02-10 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of WbpP, a Genuine UDP-N-acetylglucosamine 4-Epimerase from Pseudomonas aeruginosa: SUBSTRATE SPECIFICITY IN UDP-HEXOSE 4-EPIMERASES.
J.Biol.Chem., 279, 2004
|
|
3D3D
 
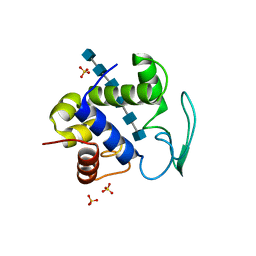 | | Bacteriophage lambda lysozyme complexed with a chitohexasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme, SULFATE ION | | Authors: | Leung, A.K.W, Berghuis, A.M. | | Deposit date: | 2008-05-09 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the lytic transglycosylase from bacteriophage lambda in complex with hexa-N-acetylchitohexaose
Biochemistry, 40, 2001
|
|
1TVE
 
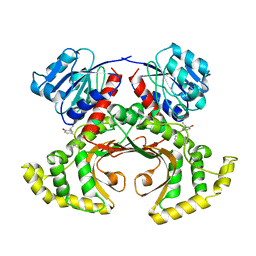 | | Homoserine Dehydrogenase in complex with 4-(4-hydroxy-3-isopropylphenylthio)-2-isopropylphenol | | Descriptor: | 4-(4-HYDROXY-3-ISOPROPYLPHENYLTHIO)-2-ISOPROPYLPHENOL, Homoserine dehydrogenase | | Authors: | Ejim, L, Mirza, I.A, Capone, C, Nazi, I, Jenkins, S, Chee, G.L, Berghuis, A.M, Wright, G.D. | | Deposit date: | 2004-06-29 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | New phenolic inhibitors of yeast homoserine dehydrogenase
Bioorg.Med.Chem., 12, 2004
|
|
1SB8
 
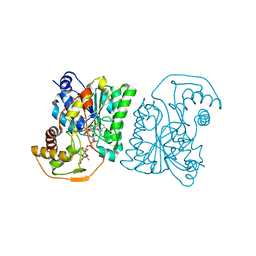 | | Crystal structure of Pseudomonas aeruginosa UDP-N-acetylglucosamine 4-epimerase complexed with UDP-N-acetylgalactosamine | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-DIPHOSPHATE-N-ACETYLGALACTOSAMINE, wbpP | | Authors: | Ishiyama, N, Creuzenet, C, Lam, J.S, Berghuis, A.M. | | Deposit date: | 2004-02-10 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of WbpP, a Genuine UDP-N-acetylglucosamine 4-Epimerase from Pseudomonas aeruginosa: SUBSTRATE SPECIFICITY IN UDP-HEXOSE 4-EPIMERASES.
J.Biol.Chem., 279, 2004
|
|
3EIG
 
 | |
6VOU
 
 | | Aminoglycoside N-2'-Acetyltransferase-Ia [AAC(2')-Ia] in complex with acetylated-plazomicin and CoA | | Descriptor: | (2S)-N-[(1R,2S,3S,4R,5S)-4-{[(2S,3R)-3-(acetylamino)-6-{[(2-hydroxyethyl)amino]methyl}-3,4-dihydro-2H-pyran-2-yl]oxy}-5-amino-2-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-3-hydroxycyclohexyl]-4-amino-2-hydroxybutanamide, (4S)-2-METHYL-2,4-PENTANEDIOL, 3,3',3''-phosphanetriyltripropanoic acid, ... | | Authors: | Bassenden, A.V, Berghuis, A.M. | | Deposit date: | 2020-01-31 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for plazomicin antibiotic action and resistance.
Commun Biol, 4, 2021
|
|
6VR3
 
 | | Aminoglycoside N-2'-Acetyltransferase-Ia [AAC(2')-Ia] in complex with acetylated-netilmicin and CoA | | Descriptor: | Aminoglycoside 2'-N-acetyltransferase, COENZYME A, N-[(2S,3R)-2-{[(1R,2S,3S,4R,6S)-6-amino-3-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-lyxopyranosyl]oxy}-4-(ethylamino) -2-hydroxycyclohexyl]oxy}-6-(aminomethyl)-3,4-dihydro-2H-pyran-3-yl]acetamide | | Authors: | Bassenden, A.V, Berghuis, A.M. | | Deposit date: | 2020-02-06 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and phylogenetic analyses of resistance to next-generation aminoglycosides conferred by AAC(2') enzymes.
Sci Rep, 11, 2021
|
|
6VR2
 
 | | Aminoglycoside N-2'-Acetyltransferase-Ia [AAC(2')-Ia] in complex with acetylated-tobramycin and CoA | | Descriptor: | (1S,2S,3R,4S,6R)-3-{[2-(acetylamino)-6-amino-2,3,6-trideoxy-alpha-D-ribo-hexopyranosyl]oxy}-4,6-diamino-2-hydroxycycloh exyl 3-amino-3-deoxy-alpha-D-glucopyranoside, ACETATE ION, Aminoglycoside 2'-N-acetyltransferase, ... | | Authors: | Bassenden, A.V, Berghuis, A.M. | | Deposit date: | 2020-02-06 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and phylogenetic analyses of resistance to next-generation aminoglycosides conferred by AAC(2') enzymes.
Sci Rep, 11, 2021
|
|
6VTA
 
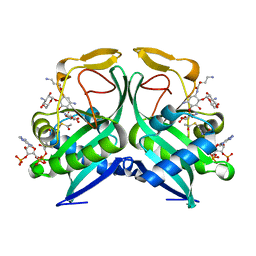 | | Aminoglycoside N-2'-Acetyltransferase-Ia [AAC(2')-Ia] in complex with amikacin and acetyl-CoA | | Descriptor: | (2S)-N-[(1R,2S,3S,4R,5S)-4-[(2R,3R,4S,5S,6R)-6-(aminomethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-5-azanyl-2-[(2S,3R,4S,5S ,6R)-4-azanyl-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-cyclohexyl]-4-azanyl-2-oxidanyl-butanamide, ACETYL COENZYME *A, Aminoglycoside 2'-N-acetyltransferase | | Authors: | Bassenden, A.V, Berghuis, A.M. | | Deposit date: | 2020-02-12 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural and phylogenetic analyses of resistance to next-generation aminoglycosides conferred by AAC(2') enzymes.
Sci Rep, 11, 2021
|
|
1J7U
 
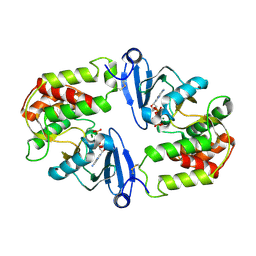 | | Crystal Structure of 3',5"-Aminoglycoside Phosphotransferase Type IIIa AMPPNP Complex | | Descriptor: | AMINOGLYCOSIDE 3'-PHOSPHOTRANSFERASE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Burk, D.L, Hon, W.C, Leung, A.K.-W, Berghuis, A.M. | | Deposit date: | 2001-05-18 | | Release date: | 2001-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analyses of nucleotide binding to an aminoglycoside phosphotransferase.
Biochemistry, 40, 2001
|
|
1J7L
 
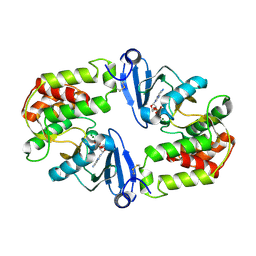 | | Crystal Structure of 3',5"-Aminoglycoside Phosphotransferase Type IIIa ADP Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AMINOGLYCOSIDE 3'-PHOSPHOTRANSFERASE, MAGNESIUM ION | | Authors: | Burk, D.L, Hon, W.C, Leung, A.K.-W, Berghuis, A.M. | | Deposit date: | 2001-05-17 | | Release date: | 2001-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analyses of nucleotide binding to an aminoglycoside phosphotransferase.
Biochemistry, 40, 2001
|
|
1J7I
 
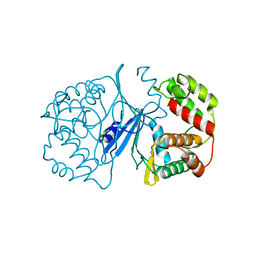 | | Crystal Structure of 3',5"-Aminoglycoside Phosphotransferase Type IIIa Apoenzyme | | Descriptor: | AMINOGLYCOSIDE 3'-PHOSPHOTRANSFERASE | | Authors: | Burk, D.L, Hon, W.C, Leung, A.K.-W, Berghuis, A.M. | | Deposit date: | 2001-05-16 | | Release date: | 2001-08-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural analyses of nucleotide binding to an aminoglycoside phosphotransferase.
Biochemistry, 40, 2001
|
|
4DEM
 
 | | Crystal structure of human FPPS in complex with YS_04_70 | | Descriptor: | Farnesyl pyrophosphate synthase, MAGNESIUM ION, [({4-[4-(propan-2-yloxy)phenyl]pyridin-2-yl}amino)methanediyl]bis(phosphonic acid) | | Authors: | Park, J, Lin, Y.-S, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2012-01-20 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design and Synthesis of Active Site Inhibitors of the Human Farnesyl Pyrophosphate Synthase: Apoptosis and Inhibition of ERK Phosphorylation in Multiple Myeloma Cells.
J.Med.Chem., 55, 2012
|
|
