5I0N
 
 | | PI4K IIalpha bound to calcium | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Phosphatidylinositol 4-kinase type 2-alpha,Lysozyme,Phosphatidylinositol 4-kinase type 2-alpha | | Authors: | Baumlova, A, Boura, E. | | Deposit date: | 2016-02-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | PI4K IIalpha bound to calcium
To Be Published
|
|
4V5E
 
 | | Insights into translational termination from the structure of RF2 bound to the ribosome | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Weixlbaumer, A, Jin, H, Neubauer, C, Voorhees, R.M, Petry, S, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-04-30 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Insights Into Translational Termination from the Structure of Rf2 Bound to the Ribosome.
Science, 322, 2008
|
|
4V5A
 
 | | Structure of the Ribosome Recycling Factor bound to the Thermus thermophilus 70S ribosome with mRNA, ASL-Phe and tRNA-fMet | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Weixlbaumer, A, Petry, S, Dunham, C.M, Selmer, M, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2007-06-28 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the ribosome recycling factor bound to the ribosome.
Nat. Struct. Mol. Biol., 14, 2007
|
|
8AC0
 
 | |
2UUC
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a Valine-ASL with cmo5U in position 34 bound to an mRNA with a GUA-codon in the A-site and paromomycin. | | Descriptor: | 16S Ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Weixlbaumer, A, Murphy, F.V, Dziergowska, A, Malkiewicz, A, Vendeix, F.A.P, Agris, P.F, Ramakrishnan, V. | | Deposit date: | 2007-03-01 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines.
Nat. Struct. Mol. Biol., 14, 2007
|
|
2UU9
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a Valine-ASL with cmo5U in position 34 bound to an mRNA with a GUG-codon in the A-site and paromomycin. | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Weixlbaumer, A, Murphy, F.V, Dziergowska, A, Malkiewicz, A, Vendeix, F.A.P, Agris, P.F, Ramakrishnan, V. | | Deposit date: | 2007-03-01 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines.
Nat. Struct. Mol. Biol., 14, 2007
|
|
2UUB
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a Valine-ASL with cmo5U in position 34 bound to an mRNA with a GUU-codon in the A-site and paromomycin. | | Descriptor: | 16S Ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Weixlbaumer, A, Murphy, F.V, Dziergowska, A, Malkiewicz, A, Vendeix, F.A.P, Agris, P.F, Ramakrishnan, V. | | Deposit date: | 2007-03-01 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism for Expanding the Decoding Capacity of Transfer Rnas by Modification of Uridines
Nat.Struct.Mol.Biol., 14, 2007
|
|
2UUA
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a Valine-ASL with cmo5U in position 34 bound to an mRNA with a GUC-codon in the A-site and paromomycin. | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Weixlbaumer, A, Murphy, F.V, Dziergowska, A, Malkiewicz, A, Vendeix, F.A.P, Agris, P.F, Ramakrishnan, V. | | Deposit date: | 2007-03-01 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines.
Nat. Struct. Mol. Biol., 14, 2007
|
|
4GZY
 
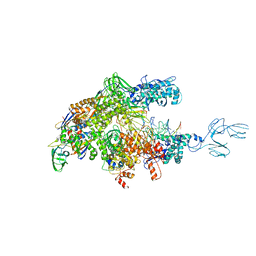 | | Crystal structures of bacterial RNA Polymerase paused elongation complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Weixlbaumer, A, Leon, K, Landick, R, Darst, S.A. | | Deposit date: | 2012-09-06 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5054 Å) | | Cite: | Structural basis of transcriptional pausing in bacteria.
Cell(Cambridge,Mass.), 152, 2013
|
|
4GZZ
 
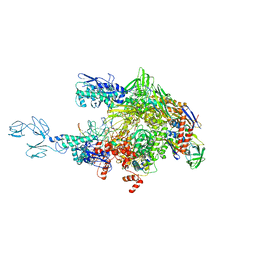 | | Crystal structures of bacterial RNA Polymerase paused elongation complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Weixlbaumer, A, Leon, K, Landick, R, Darst, S.A. | | Deposit date: | 2012-09-06 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.2927 Å) | | Cite: | Structural basis of transcriptional pausing in bacteria.
Cell(Cambridge,Mass.), 152, 2013
|
|
4PLA
 
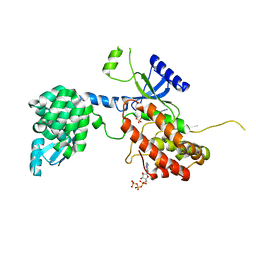 | |
5EUT
 
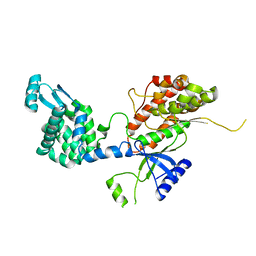 | |
7TN9
 
 | | Structure of the Inmazeb cocktail and resistance to escape against Ebola virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, GP2, ... | | Authors: | Rayaprolu, V, Fulton, B, Rafique, A, Arturo, E, Williams, D, Hariharan, C, Callaway, H, Parvate, A, Schendel, S.L, Parekh, D, Hui, S, Shaffer, K, Pascal, K.E, Wloga, E, Giordano, S, Copin, R, Franklin, M, Boytz, R.M, Donahue, C, Davey, R, Baum, A, Kyratsous, C.A, Saphire, E.O. | | Deposit date: | 2022-01-20 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the Inmazeb cocktail and resistance to Ebola virus escape.
Cell Host Microbe, 31, 2023
|
|
5VTE
 
 | |
5AH1
 
 | | Structure of EstA from Clostridium botulinum | | Descriptor: | POTASSIUM ION, TRIACYLGLYCEROL LIPASE, ZINC ION | | Authors: | Pairitsch, A, Lyskowski, A, Hromic, A, Steinkellner, G, Gruber, K, Perz, V, Baumschlager, A, Bleymaier, K, Zitzenbacher, S, Sinkel, C, Kueper, U, Ribitsch, D, Guebitz, G.M. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Hydrolysis of Synthetic Polyesters by Clostridium Botulinum Esterases.
Biotechnol.Bioeng., 113, 2016
|
|
4V5D
 
 | | Structure of the Thermus thermophilus 70S ribosome in complex with mRNA, paromomycin, acylated A- and P-site tRNAs, and E-site tRNA. | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Voorhees, R.M, Weixlbaumer, A, Loakes, D, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-03-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome.
Nat. Struct. Mol. Biol., 16, 2009
|
|
4V5C
 
 | | Structure of the Thermus thermophilus 70S ribosome in complex with mRNA, paromomycin, acylated A-site tRNA, deacylated P-site tRNA, and E-site tRNA. | | Descriptor: | 16S ribosomal RNA, 23S Ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Voorhees, R.M, Weixlbaumer, A, Loakes, D, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-03-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Insights Into Substrate Stabilization from Snapshots of the Peptidyl Transferase Center of the Intact 70S Ribosome
Nat.Struct.Mol.Biol., 16, 2009
|
|
4V51
 
 | | Structure of the Thermus thermophilus 70S ribosome complexed with mRNA, tRNA and paromomycin | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Selmer, M, Dunham, C.M, Murphy, F.V, Weixlbaumer, A, Petry, S, Weir, J.R, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2006-07-31 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the 70S ribosome complexed with mRNA and tRNA.
Science, 313, 2006
|
|
4V5F
 
 | | The structure of the ribosome with elongation factor G trapped in the post-translocational state | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, Y.-G, Selmer, M, Dunham, C.M, Weixlbaumer, A, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-09-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The structure of the ribosome with elongation factor G trapped in the posttranslocational state.
Science, 326, 2009
|
|
6RI7
 
 | | Cryo-EM structure of E. coli RNA polymerase elongation complex bound to GreB transcription factor | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
6RH3
 
 | | Cryo-EM structure of E. coli RNA polymerase elongation complex bound to CTP substrate | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-18 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
5TXK
 
 | | CRYSTAL STRUCTURE OF USP35 C450S IN COMPLEX WITH UBIQUITIN | | Descriptor: | 1,2-ETHANEDIOL, Polyubiquitin-B, SULFATE ION, ... | | Authors: | Bader, G, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2016-11-17 | | Release date: | 2018-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Expansion of DUB functionality generated by alternative isoforms - USP35, a case study.
J. Cell. Sci., 131, 2018
|
|
5MW2
 
 | | CRYSTAL STRUCTURE OF BCL-6 BTB-domain with BI-3802 | | Descriptor: | 2-[6-[[5-chloranyl-2-[(3~{S},5~{R})-3,5-dimethylpiperidin-1-yl]pyrimidin-4-yl]amino]-1-methyl-2-oxidanylidene-quinolin-3-yl]oxy-~{N}-methyl-ethanamide, B-cell lymphoma 6 protein | | Authors: | Bader, G, Flotzinger, G, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2017-01-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6.
Cell Rep, 20, 2017
|
|
8AHC
 
 | | Crystal structure of the BRD9 bromodomain with BI-7189 | | Descriptor: | Bromodomain-containing protein 9, [2,6-dimethoxy-4-(1,2,5-trimethyl-6-oxidanylidene-pyridin-3-yl)phenyl]methyl-dimethyl-azanium | | Authors: | Bader, G, Boettcher, J, Weiss-Puxbaum, A. | | Deposit date: | 2022-07-21 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | Discovery of a Chemical Probe to Study Implications of BPTF Bromodomain Inhibition in Cellular and in vivo Experiments.
Chemmedchem, 18, 2023
|
|
5MWD
 
 | | Crystal structure of the BCL6 BTB-domain with compound 2 | | Descriptor: | 5-[[5-chloranyl-2-(3,5-dimethylpyrazol-1-yl)pyrimidin-4-yl]amino]-1,3-dihydroindol-2-one, B-cell lymphoma 6 protein | | Authors: | Bader, G, Flotzinger, G, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2017-01-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6.
Cell Rep, 20, 2017
|
|
