1RX0
 
 | | Crystal structure of isobutyryl-CoA dehydrogenase complexed with substrate/ligand. | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Acyl-CoA dehydrogenase family member 8, ... | | Authors: | Battaile, K.P, Nguyen, T.V, Vockley, J, Kim, J.J. | | Deposit date: | 2003-12-18 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structures of Isobutyryl-CoA Dehydrogenase and Enzyme-Product Complex: COMPARISON WITH ISOVALERYL- AND SHORT-CHAIN ACYL-COA DEHYDROGENASES.
J.Biol.Chem., 279, 2004
|
|
5KEF
 
 | | Structure of hypothetical Staphylococcus protein SA0856 with zinc | | Descriptor: | ACETATE ION, PhnB protein, ZINC ION | | Authors: | Battaile, K.P, Chirgadze, Y.N, Lam, R, Chan, T, Mihajlovic, V, Romanov, V, Pai, E, Mendez, V, Chirgadze, N.Y. | | Deposit date: | 2016-06-09 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structure of Staphylococcus aureus Zn-glyoxalase I: new subfamily of glyoxalase I family.
J. Biomol. Struct. Dyn., 36, 2018
|
|
2BZG
 
 | | Crystal structure of thiopurine S-methyltransferase. | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYL-L-HOMOCYSTEINE, THIOPURINE S-METHYLTRANSFERASE | | Authors: | Battaile, K.P, Wu, H, Zeng, H, Loppnau, P, Dong, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-17 | | Release date: | 2005-08-25 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Basis of Allele Variation of Human Thiopurine-S-Methyltransferase.
Proteins, 67, 2007
|
|
1JQI
 
 | | Crystal Structure of Rat Short Chain Acyl-CoA Dehydrogenase Complexed With Acetoacetyl-CoA | | Descriptor: | ACETOACETYL-COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, short chain acyl-CoA dehydrogenase | | Authors: | Battaile, K.P, Molin-Case, J, Paschke, R, Wang, M, Bennett, D, Vockley, J, Kim, J.-J.P. | | Deposit date: | 2001-08-07 | | Release date: | 2002-02-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of rat short chain acyl-CoA dehydrogenase complexed with acetoacetyl-CoA: comparison with other acyl-CoA dehydrogenases.
J.Biol.Chem., 277, 2002
|
|
7KEY
 
 | | Protein Tyrosine Phosphatase 1B, Apo | | Descriptor: | ACETATE ION, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Battaile, K.P, Chirgadze, Y, Ruzanov, M, Romanov, V, Lam, K, Gordon, R, Lin, A, Lam, R, Pai, E, Chirgadze, N. | | Deposit date: | 2020-10-13 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | Signal transfer in human protein tyrosine phosphatase PTP1B from allosteric inhibitor P00058.
J.Biomol.Struct.Dyn., 2021
|
|
7KLX
 
 | | Protein Tyrosine Phosphatase 1B with inhibitor | | Descriptor: | 2-(2,5-dimethyl-1H-pyrrol-1-yl)-5-hydroxybenzoic acid, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Battaile, K.P, Chirgadze, Y, Ruzanov, M, Romanov, V, Lam, K, Gordon, R, Lin, A, Lam, R, Pai, E, Chirgadze, N. | | Deposit date: | 2020-11-01 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | Signal transfer in human protein tyrosine phosphatase PTP1B from allosteric inhibitor P00058.
J.Biomol.Struct.Dyn., 2021
|
|
4PAV
 
 | | Structure of hypothetical protein SA1046 from S. aureus. | | Descriptor: | Glyoxalase family protein | | Authors: | Battaile, K.P, Mulichak, A, Lam, R, Lam, K, Soloveychik, M, Romanov, V, Jones, K, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-04-10 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of hypothetical protein SA1046 from S. aureus.
To Be Published
|
|
4PAW
 
 | | Structure of hypothetical protein HP1257. | | Descriptor: | Orotate phosphoribosyltransferase, PHOSPHATE ION, THIOCYANATE ION, ... | | Authors: | Battaile, K.P, Lam, R, Lam, K, Romanov, V, Jones, K, Soloveychik, M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-04-10 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure of hypothetical protein HP1257.
To Be Published
|
|
4PAU
 
 | | Hypothetical protein SA1058 from S. aureus. | | Descriptor: | CHLORIDE ION, Nitrogen regulatory protein A | | Authors: | Battaile, K.P, Wu-Brown, J, Romanov, V, Jones, K, Lam, R, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-04-10 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hypothetical protein SA1058 from S. aureus.
To Be Published
|
|
4P13
 
 | |
3UR9
 
 | | 1.65A resolution structure of Norwalk Virus Protease Containing a covalently bound dipeptidyl inhibitor | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like protease, CHLORIDE ION | | Authors: | Lovell, S, Battaile, K.P, Kim, Y, Tiew, K.C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.O. | | Deposit date: | 2011-11-21 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Antivirals against 3C or 3C-Like Proteases of Picornaviruses, Noroviruses, and Coronaviruses.
J.Virol., 86, 2012
|
|
3UR6
 
 | | 1.5A resolution structure of apo Norwalk Virus Protease | | Descriptor: | 3C-like protease | | Authors: | Lovell, S, Battaile, K.P, Kim, Y, Tiew, K.C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.O. | | Deposit date: | 2011-11-21 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Broad-Spectrum Antivirals against 3C or 3C-Like Proteases of Picornaviruses, Noroviruses, and Coronaviruses.
J.Virol., 86, 2012
|
|
7RT0
 
 | |
5D8P
 
 | | 2.35A resolution structure of iron bound BfrB (wild-type, C2221 form) from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, FE (II) ION, Ferroxidase, ... | | Authors: | Lovell, S, Battaile, K.P, Wang, Y, Yao, H, Rivera, M. | | Deposit date: | 2015-08-17 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Characterization of the Bacterioferritin/Bacterioferritin Associated Ferredoxin Protein-Protein Interaction in Solution and Determination of Binding Energy Hot Spots.
Biochemistry, 54, 2015
|
|
5D8X
 
 | | 1.50A resolution structure of BfrB (L68A E81A) from Pseudomonas aeruginosa | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Ferroxidase, ... | | Authors: | Lovell, S, Battaile, K.P, Wang, Y, Yao, H, Rivera, M. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization of the Bacterioferritin/Bacterioferritin Associated Ferredoxin Protein-Protein Interaction in Solution and Determination of Binding Energy Hot Spots.
Biochemistry, 54, 2015
|
|
4FA0
 
 | | Crystal structure of human AdPLA to 2.65 A resolution | | Descriptor: | Group XVI phospholipase A1/A2 | | Authors: | Lovell, S, Battaile, K.P, Addington, L, Zhang, N, Rao, J.L.U.M, Moise, A.R. | | Deposit date: | 2012-05-21 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure/Function relationships of adipose phospholipase A2 containing a cys-his-his catalytic triad.
J.Biol.Chem., 287, 2012
|
|
4FP7
 
 | |
4O8I
 
 | | 1.45A resolution structure of PEG 400 Bound Cyclophilin D | | Descriptor: | PENTAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Lovell, S, Valasani, K.R, Battaile, K.P, Wang, C, Yan, S.S. | | Deposit date: | 2013-12-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution crystal structures of two crystal forms of human cyclophilin D in complex with PEG 400 molecules.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4O8H
 
 | | 0.85A resolution structure of PEG 400 Bound Cyclophilin D | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Lovell, S, Valasani, K.R, Battaile, K.P, Wang, C, Yan, S.S. | | Deposit date: | 2013-12-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | High-resolution crystal structures of two crystal forms of human cyclophilin D in complex with PEG 400 molecules.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
5DG6
 
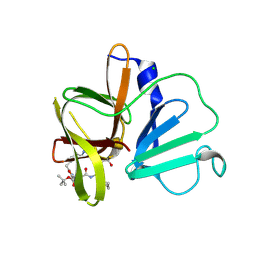 | | 2.35A resolution structure of Norovirus 3CL protease in complex an oxadiazole-based, cell permeable macrocyclic (21-mer) inhibitor | | Descriptor: | 3C-LIKE PROTEASE, CHLORIDE ION, tert-butyl [(4S,7S,10S)-7-(cyclohexylmethyl)-10-(hydroxymethyl)-5,8,13-trioxo-23-oxa-6,9,14,21,22-pentaazabicyclo[18.2.1]tricosa-1(22),20-dien-4-yl]carbamate | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Damalanka, V.C, Kim, Y, Alliston, K.R, Weerawarna, P.M, Kankanamalage, A.C.G, Lushington, G.H, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2015-08-27 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Oxadiazole-Based Cell Permeable Macrocyclic Transition State Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 59, 2016
|
|
5D8R
 
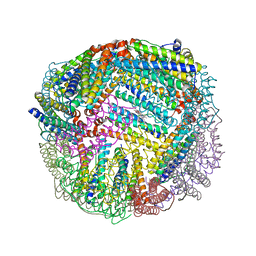 | | 2.50A resolution structure of BfrB (E81A) from Pseudomonas aeruginosa | | Descriptor: | Ferroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Battaile, K.P, Wang, Y, Yao, H, Rivera, M. | | Deposit date: | 2015-08-17 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of the Bacterioferritin/Bacterioferritin Associated Ferredoxin Protein-Protein Interaction in Solution and Determination of Binding Energy Hot Spots.
Biochemistry, 54, 2015
|
|
5D8S
 
 | | 2.55A resolution structure of BfrB (E85A) from Pseudomonas aeruginosa | | Descriptor: | Ferroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Battaile, K.P, Wang, Y, Yao, H, Rivera, M. | | Deposit date: | 2015-08-17 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Characterization of the Bacterioferritin/Bacterioferritin Associated Ferredoxin Protein-Protein Interaction in Solution and Determination of Binding Energy Hot Spots.
Biochemistry, 54, 2015
|
|
4O06
 
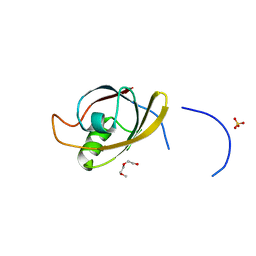 | | 1.15A Resolution Structure of the Proteasome Assembly Chaperone Nas2 PDZ Domain | | Descriptor: | Probable 26S proteasome regulatory subunit p27, SULFATE ION, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Singh, C.R, Chowdhury, W.Q, Geanes, E, Roelofs, J. | | Deposit date: | 2013-12-13 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | 1.15 angstrom resolution structure of the proteasome-assembly chaperone Nas2 PDZ domain.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4O6Q
 
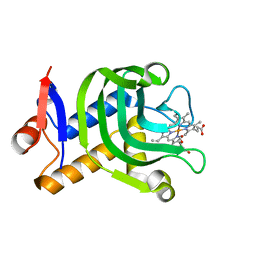 | | 0.95A resolution structure of the hemophore HasA from Pseudomonas aeruginosa (Y75A mutant) | | Descriptor: | FORMIC ACID, HasAp, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Kumar, R, Battaile, K.P, Matsumura, H, Yao, H, Rodriguez, J.C, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Replacing the Axial Ligand Tyrosine 75 or Its Hydrogen Bond Partner Histidine 83 Minimally Affects Hemin Acquisition by the Hemophore HasAp from Pseudomonas aeruginosa.
Biochemistry, 53, 2014
|
|
6XMK
 
 | | 1.70 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 7j | | Descriptor: | (1S,2S)-2-[(N-{[(4,4-difluorocyclohexyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
