1RPM
 
 | |
1H6G
 
 | | alpha-catenin M-domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ALPHA-1 CATENIN, CALCIUM ION, ... | | Authors: | Yang, J, Dokurno, P, Tonks, N.K, Barford, D. | | Deposit date: | 2001-06-14 | | Release date: | 2001-08-07 | | Last modified: | 2016-02-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the M-Fragment of Alpha-Catenin: Implications for Modulation of Cell Adhesion.
Embo J., 20, 2001
|
|
4AEZ
 
 | | Crystal Structure of Mitotic Checkpoint Complex | | Descriptor: | MITOTIC SPINDLE CHECKPOINT COMPONENT MAD2, MITOTIC SPINDLE CHECKPOINT COMPONENT MAD3, WD REPEAT-CONTAINING PROTEIN SLP1 | | Authors: | Kulkarni, K.A, Chao, W.C.H, Zhang, Z, Barford, D. | | Deposit date: | 2012-01-13 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Mitotic Checkpoint Complex
Nature, 484, 2012
|
|
4BH6
 
 | | Insights into degron recognition by APC coactivators from the structure of an Acm1-Cdh1 complex | | Descriptor: | APC/C ACTIVATOR PROTEIN CDH1, APC/C-CDH1 MODULATOR 1 | | Authors: | He, J, Chao, W.C.H, Zhang, Z, Yang, J, Cronin, N, Barford, D. | | Deposit date: | 2013-03-29 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights Into Degron Recognition by Apc/C Coactivators from the Structure of an Acm1-Cdh1 Complex.
Mol.Cell, 50, 2013
|
|
3ZJE
 
 | | A20 OTU domain in reversibly oxidised (SOH) state | | Descriptor: | 1,2-ETHANEDIOL, A20P50, CHLORIDE ION | | Authors: | Kulathu, Y, Garcia, F.J, Mevissen, T.E.T, Busch, M, Arnaudo, N, Carroll, K.S, Barford, D, Komander, D. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Regulation of A20 and Other Otu Deubiquitinases by Reversible Oxidation
Nat.Commun., 4, 2013
|
|
3ZJD
 
 | | A20 OTU domain in reduced, active state at 1.87 A resolution | | Descriptor: | 1,2-ETHANEDIOL, A20P50, CHLORIDE ION | | Authors: | Kulathu, Y, Garcia, F.J, Mevissen, T.E.T, Busch, M, Arnaudo, N, Carroll, K.S, Barford, D, Komander, D. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Regulation of A20 and Other Otu Deubiquitinases by Reversible Oxidation
Nat.Commun., 4, 2013
|
|
3ZJF
 
 | | A20 OTU domain with irreversibly oxidised Cys103 from 270 min H2O2 soak. | | Descriptor: | A20P50, CHLORIDE ION | | Authors: | Kulathu, Y, Garcia, F.J, Mevissen, T.E.T, Busch, M, Arnaudo, N, Carroll, K.S, Barford, D, Komander, D. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Regulation of A20 and Other Otu Deubiquitinases by Reversible Oxidation
Nat.Commun., 4, 2013
|
|
3ZJG
 
 | | A20 OTU domain with irreversibly oxidised Cys103 from 60 min H2O2 soak. | | Descriptor: | CHLORIDE ION, TUMOR NECROSIS FACTOR ALPHA-INDUCED PROTEIN 3 | | Authors: | Kulathu, Y, Garcia, F.J, Mevissen, T.E.T, Busch, M, Arnaudo, N, Carroll, K.S, Barford, D, Komander, D. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Regulation of A20 and Other Otu Deubiquitinases by Reversible Oxidation
Nat.Commun., 4, 2013
|
|
4ADY
 
 | | Crystal structure of 26S proteasome subunit Rpn2 | | Descriptor: | 26S PROTEASOME REGULATORY SUBUNIT RPN2 | | Authors: | Kulkarni, K, He, J, Da Fonseca, P.C.A, Krutauz, D, Glickman, M.H, Barford, D, Morris, E.P. | | Deposit date: | 2012-01-04 | | Release date: | 2012-03-14 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of the 26S Proteasome Subunit Rpn2 Reveals its Pc Repeat Domain as a Closed Toroid of Two Concentric Alpha-Helical Rings
Structure, 20, 2012
|
|
1FPZ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF KINASE ASSOCIATED PHOSPHATASE (KAP) WITH A SUBSTITUTION OF THE CATALYTIC SITE CYSTEINE (CYS140) TO A SERINE | | Descriptor: | CYCLIN-DEPENDENT KINASE INHIBITOR 3, SULFATE ION | | Authors: | Song, H, Hanlon, N, Brown, N.R, Noble, M.E.M, Johnson, L.N, Barford, D. | | Deposit date: | 2000-09-01 | | Release date: | 2001-05-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphoprotein-protein interactions revealed by the crystal structure of kinase-associated phosphatase in complex with phosphoCDK2.
Mol.Cell, 7, 2001
|
|
4A2N
 
 | | Crystal Structure of Ma-ICMT | | Descriptor: | CARDIOLIPIN, ISOPRENYLCYSTEINE CARBOXYL METHYLTRANSFERASE, PALMITIC ACID, ... | | Authors: | Yang, J, Kulkarni, K, Manolaridis, I, Zhang, Z, Dodd, R.B, Mas-Droux, C, Barford, D. | | Deposit date: | 2011-09-27 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mechanism of Isoprenylcysteine Carboxyl Methylation from the Crystal Structure of the Integral Membrane Methyltransferase Icmt.
Mol.Cell, 44, 2011
|
|
1O6K
 
 | | Structure of activated form of PKB kinase domain S474D with GSK3 peptide and AMP-PNP | | Descriptor: | GLYCOGEN SYNTHASE KINASE-3 BETA, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Yang, J, Cron, P, Good, V.M, Thompson, V, Hemmings, B.A, Barford, D. | | Deposit date: | 2002-10-08 | | Release date: | 2002-11-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of an Activated Akt/Protein Kinase B Ternary Complex with Gsk-3 Peptide and AMP-Pnp
Nat.Struct.Biol., 9, 2002
|
|
1O6L
 
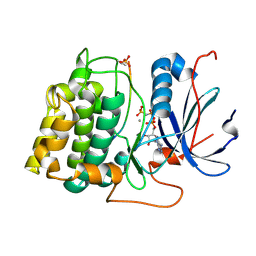 | | Crystal structure of an activated Akt/protein kinase B (PKB-PIF chimera) ternary complex with AMP-PNP and GSK3 peptide | | Descriptor: | GLYCOGEN SYNTHASE KINASE-3 BETA, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Yang, J, Cron, P, Good, V.M, Thompson, V, Hemmings, B.A, Barford, D. | | Deposit date: | 2002-10-08 | | Release date: | 2002-11-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of an Activated Akt/Protein Kinase B Ternary Complex with Gsk-3 Peptide and AMP-Pnp
Nat.Struct.Biol., 9, 2002
|
|
1OHE
 
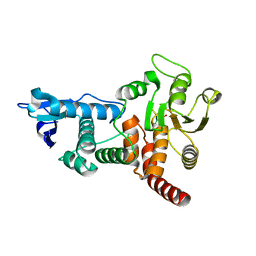 | | Structure of cdc14b phosphatase with a peptide ligand | | Descriptor: | CDC14B2 PHOSPHATASE, PEPTIDE LIGAND | | Authors: | Gray, C.H, Good, V.M, Tonks, N.K, Barford, D. | | Deposit date: | 2003-05-24 | | Release date: | 2003-07-24 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of the Cell Cycle Protein Cdc14 Reveals a Proline-Directed Protein Phosphatase
Embo J., 22, 2003
|
|
1FQ1
 
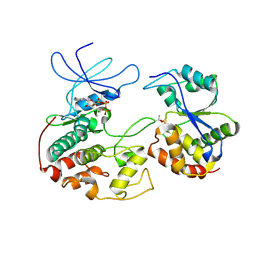 | | CRYSTAL STRUCTURE OF KINASE ASSOCIATED PHOSPHATASE (KAP) IN COMPLEX WITH PHOSPHO-CDK2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION PROTEIN KINASE 2, CYCLIN-DEPENDENT KINASE INHIBITOR 3, ... | | Authors: | Song, H, Hanlon, N, Brown, N.R, Noble, M.E.M, Johnson, L.N, Barford, D. | | Deposit date: | 2000-09-01 | | Release date: | 2001-05-09 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phosphoprotein-protein interactions revealed by the crystal structure of kinase-associated phosphatase in complex with phosphoCDK2.
Mol.Cell, 7, 2001
|
|
1G1H
 
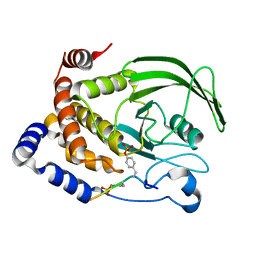 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH A BIS-PHOSPHORYLATED PEPTIDE (ETD(PTR)(PTR)RKGGKGLL) FROM THE INSULIN RECEPTOR KINASE | | Descriptor: | BI-PHOSPHORYLATED PEPTIDE FROM THE INSULIN RECEPTOR KINASE, PROTEIN TYROSINE PHOSPHATASE 1B | | Authors: | Salmeen, A, Andersen, J.N, Myers, M.P, Tonks, N.K, Barford, D. | | Deposit date: | 2000-10-11 | | Release date: | 2001-01-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B.
Mol.Cell, 6, 2000
|
|
1G1G
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH A MONO-PHOSPHORYLATED PEPTIDE (ETDY(PTR)RKGGKGLL) FROM THE INSULIN RECEPTOR KINASE | | Descriptor: | MONO-PHOSPHORYLATED PEPTIDE FROM THE INSULIN RECEPTOR KINASE, PROTEIN TYROSINE PHOSPHATASE 1B | | Authors: | Salmeen, A, Andersen, J.N, Myers, M.P, Tonks, N.K, Barford, D. | | Deposit date: | 2000-10-11 | | Release date: | 2001-01-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B.
Mol.Cell, 6, 2000
|
|
1G1F
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH A TRI-PHOSPHORYLATED PEPTIDE (RDI(PTR)ETD(PTR)(PTR)RK) FROM THE INSULIN RECEPTOR KINASE | | Descriptor: | PROTEIN TYROSINE PHOSPHATASE 1B, TRI-PHOSPHORYLATED PEPTIDE FROM THE INSULIN RECEPTOR KINASE | | Authors: | Salmeen, A, Andersen, J.N, Myers, M.P, Tonks, N.K, Barford, D. | | Deposit date: | 2000-10-11 | | Release date: | 2001-01-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B.
Mol.Cell, 6, 2000
|
|
3ZN3
 
 | | N-terminal domain of S. pombe Cdc23 APC subunit | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 8, MERCURY (II) ION | | Authors: | Zhang, Z, Yang, J, Conin, N, Kulkarni, K, Barford, D. | | Deposit date: | 2013-02-13 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Four Canonical Tpr Subunits of Human Apc/C Form Related Homo-Dimeric Structures and Stack in Parallel to Form a Tpr Suprahelix
J.Mol.Biol., 425, 2013
|
|
1OHC
 
 | |
2IZY
 
 | | Molecular Basis of AKAP Specificity for PKA Regulatory Subunits | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE REGULATORY SUBUNIT II | | Authors: | Gold, M.G, Lygren, B, Dokurno, P, Hoshi, N, McConnachie, G, Tasken, K, Carlson, C.R, Scott, J.D, Barford, D. | | Deposit date: | 2006-07-27 | | Release date: | 2006-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Basis of Akap Specificity for Pka Regululatory Subunits
Mol.Cell, 24, 2006
|
|
2JF5
 
 | | crystal structure of Lys63-linked di-ubiquitin | | Descriptor: | CADMIUM ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Komander, D, Odenwaelder, P, Barford, D. | | Deposit date: | 2007-01-26 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Discrimination of Structurally Equivalent Lys 63-Linked and Linear Polyubiquitin Chains.
Embo Rep., 10, 2009
|
|
1OHD
 
 | | structure of cdc14 in complex with tungstate | | Descriptor: | CDC14B2 PHOSPHATASE, TUNGSTATE(VI)ION | | Authors: | Gray, C.H, Good, V.M, Tonks, N.K, Barford, D. | | Deposit date: | 2003-05-24 | | Release date: | 2003-07-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structure of the Cell Cycle Protein Cdc14 Reveals a Proline-Directed Protein Phosphatase
Embo J., 22, 2003
|
|
2IZX
 
 | | Molecular Basis of AKAP Specificity for PKA Regulatory Subunits | | Descriptor: | AKAP-IS, CAMP-DEPENDENT PROTEIN KINASE TYPE II-ALPHA REGULATORY SUBUNIT, DITHIANE DIOL | | Authors: | Gold, M.G, Lygren, B, Dokurno, P, Hoshi, N, McConnachie, G, Tasken, K, Carlson, C.R, Scott, J.D, Barford, D. | | Deposit date: | 2006-07-27 | | Release date: | 2006-11-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular Basis of Akap Specificity for Pka Regulatory Subunits.
Mol.Cell, 24, 2006
|
|
2W9N
 
 | | crystal structure of linear di-ubiquitin | | Descriptor: | CHLORIDE ION, UBIQUITIN, ZINC ION | | Authors: | Komander, D, Reyes-Turcu, F, Wilkinson, K.D, Barford, D. | | Deposit date: | 2009-01-27 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular Discrimination of Structurally Equivalent Lys 63-Linked and Linear Polyubiquitin Chains.
Embo Rep., 10, 2009
|
|
