3UD6
 
 | | Structural analyses of covalent enzyme-substrate analogue complexes reveal strengths and limitations of de novo enzyme design | | Descriptor: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, RETRO-ALDOLASE, SULFATE ION | | Authors: | Baker, D, Stoddard, B.L, Althoff, E.A, Wang, L, Jiang, L, Moody, J, Bolduc, J, Lassila, J, Hilvert, D. | | Deposit date: | 2011-10-27 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
1VJQ
 
 | |
6DM9
 
 | | DHD15_extended | | Descriptor: | DHD15_extended_A, DHD15_extended_B, SULFATE ION | | Authors: | Bick, M.J, Chen, Z, Baker, D. | | Deposit date: | 2018-06-04 | | Release date: | 2018-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
7MWQ
 
 | | Structure of De Novo designed beta sheet heterodimer LHD29A53/B53 | | Descriptor: | LHD29A53, LHD29B53 | | Authors: | Bera, A.K, Sahtoe, D.D, Kang, A, Praetorius, F, Baker, D. | | Deposit date: | 2021-05-17 | | Release date: | 2022-01-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Reconfigurable asymmetric protein assemblies through implicit negative design.
Science, 375, 2022
|
|
7MWR
 
 | | Structure of De Novo designed beta sheet heterodimer LHD101A53/B4 | | Descriptor: | LHD101A54, LHD101B4, MALONATE ION | | Authors: | Bera, A.K, Sahtoe, D.D, Kang, A, Praetorius, F, Baker, D. | | Deposit date: | 2021-05-17 | | Release date: | 2022-01-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reconfigurable asymmetric protein assemblies through implicit negative design.
Science, 375, 2022
|
|
6DLM
 
 | | DHD127 | | Descriptor: | DHD127_A, DHD127_B | | Authors: | Bick, M.J, Chen, Z, Baker, D. | | Deposit date: | 2018-06-01 | | Release date: | 2018-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
6DMA
 
 | | DHD15_closed | | Descriptor: | DHD15_closed_A, DHD15_closed_B | | Authors: | Bick, M.J, Chen, Z, Baker, D. | | Deposit date: | 2018-06-04 | | Release date: | 2018-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.363 Å) | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
6DKM
 
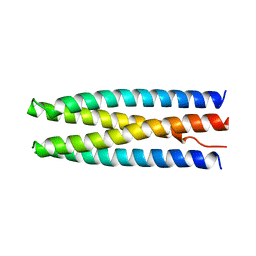 | | DHD131 | | Descriptor: | DHD131_A, DHD131_B | | Authors: | Bick, M.J, Chen, Z, Baker, D. | | Deposit date: | 2018-05-29 | | Release date: | 2018-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
5HRY
 
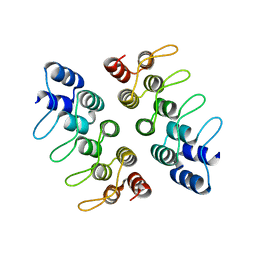 | | Computationally Designed Cyclic Dimer ank3C2_1 | | Descriptor: | ank3C2_1 | | Authors: | Cascio, D, McNamara, D.E, Fallas, J.A, Baker, D, Yeates, T.O. | | Deposit date: | 2016-01-24 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
3UYC
 
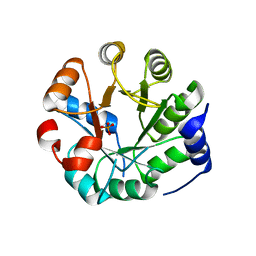 | | Designed protein KE59 R8_2/7A | | Descriptor: | Kemp eliminase KE59 R8_2/7A, PHOSPHATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-06 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FFD
 
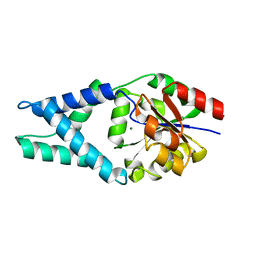 | | Crystal structure of engineered protein. northeast structural genomics consortium target or48 | | Descriptor: | MAGNESIUM ION, PF00702 DOMAIN PROTEIN | | Authors: | Seetharaman, J, Lew, S, Nivon, L, Baker, D, Bjelic, S, Ciccosanti, C, Sahdev, S, Xiao, R, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-31 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of engineered protein. northeast structural genomics consortium target or48
To be Published
|
|
3UXD
 
 | | Designed protein KE59 R1 7/10H with dichlorobenzotriazole (DBT) | | Descriptor: | 5,7-dichloro-1H-benzotriazole, Kemp eliminase KE59 R1 7/10H, PHOSPHATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-05 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UY7
 
 | | Designed protein KE59 R1 7/10H with G130S mutation | | Descriptor: | Kemp eliminase KE59 R1 7/10H, SODIUM ION, SULFATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-06 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4ZK7
 
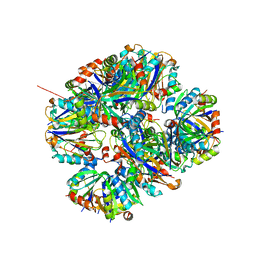 | | Crystal structure of rescued two-component self-assembling tetrahedral cage T33-31 | | Descriptor: | Chorismate mutase, Divalent-cation tolerance protein CutA | | Authors: | Liu, Y, Cascio, D, Sawaya, M.R, Bale, J, Collazo, M.J, Park, R, King, N, Baker, D, Yeates, T. | | Deposit date: | 2015-04-30 | | Release date: | 2015-07-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a designed tetrahedral protein assembly variant engineered to have improved soluble expression.
Protein Sci., 24, 2015
|
|
3LEV
 
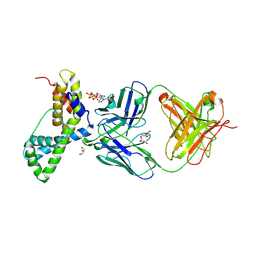 | | HIV-1 antibody 2F5 in complex with epitope scaffold ES2 | | Descriptor: | 2F5 ANTIBODY HEAVY CHAIN, 2F5 ANTIBODY LIGHT CHAIN, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ofek, G, Guenaga, F.J, Schief, W.R, Skinner, J, Baker, D, Wyatt, R, Kwong, P.D. | | Deposit date: | 2010-01-15 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Elicitation of structure-specific antibodies by epitope scaffolds.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3LES
 
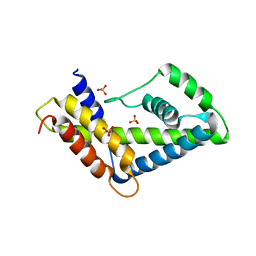 | | 2F5 Epitope scaffold ES2 | | Descriptor: | RNA polymerase sigma factor, SULFATE ION | | Authors: | Ofek, G, Guenaga, F.J, Schief, W.R, Skinner, J, Wyatt, R, Baker, D, Kwong, P.D. | | Deposit date: | 2010-01-15 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Elicitation of structure-specific antibodies by epitope scaffolds.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3LEY
 
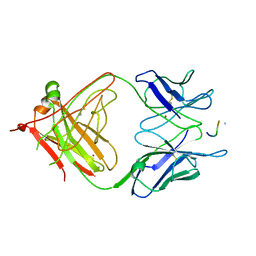 | | 2F5 Epitope scaffold elicited anti-HIV-1 monoclonal antibody 6a7 in complex with HIV-1 GP41 | | Descriptor: | 6a7 Antibody Heavy Chain, 6a7 Antibody Light Chain, Envelope glycoprotein gp41, ... | | Authors: | Ofek, G, Guenaga, F.J, Schief, W.R, Skinner, J, Baker, D, Wyatt, R, Kwong, P.D. | | Deposit date: | 2010-01-15 | | Release date: | 2010-09-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Elicitation of structure-specific antibodies by epitope scaffolds.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5UP1
 
 | | Solution structure of the de novo mini protein EEHEE_rd3_1049 | | Descriptor: | EEHEE_rd3_1049 | | Authors: | Houliston, S, Rocklin, G.J, Lemak, A, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | Deposit date: | 2017-02-01 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
6WRX
 
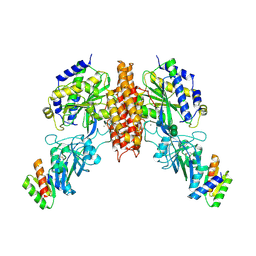 | | Crystal structure of computationally designed protein 2DS25.1 in complex with the human Transferrin receptor ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Abraham, J, Coscia, A, Olal, D, Sahtoe, D.D, Baker, D, Clark, L. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Transferrin receptor targeting by de novo sheet extension.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6WRW
 
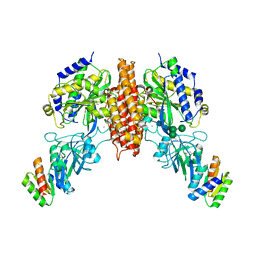 | | Crystal structure of computationally designed protein 2DS25.5 in complex with the human Transferrin receptor ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Computationally designed protein 2DS25.5, ... | | Authors: | Abraham, J, Coscia, A, Olal, D, Sahtoe, D.D, Baker, D, Clark, L. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Transferrin receptor targeting by de novo sheet extension.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6WRV
 
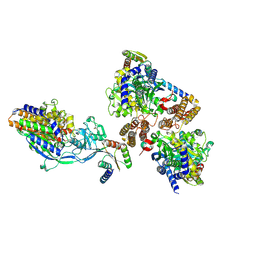 | | Crystal structure of computationally designed protein 3DS18 in complex with the human Transferrin receptor ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Computationally designed protein 3DS18, ... | | Authors: | Abraham, J, Baker, D, Sahtoe, D.D, Coscia, A, Clark, L, Olal, D. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Transferrin receptor targeting by de novo sheet extension.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RMY
 
 | | De Novo designed tunable protein pockets, D_3-337 | | Descriptor: | De Novo designed tunable homodimer, D_3-337 | | Authors: | Bera, A.K, Hicks, D.R, Kang, A, Sankaran, B, Baker, D. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | De novo design of protein homodimers containing tunable symmetric protein pockets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RMX
 
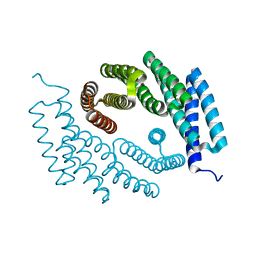 | | Structure of De Novo designed tunable symmetric protein pockets | | Descriptor: | Tunable symmetric protein, D_3_212 | | Authors: | Bera, A.K, Hicks, D.R, Kang, A, Sankaran, B, Baker, D. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | De novo design of protein homodimers containing tunable symmetric protein pockets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7M5T
 
 | |
6WHZ
 
 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | Histone deacetylase 2, SODIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
