8DYM
 
 | | Aspartimidylated Graspetide Amycolimiditide | | Descriptor: | ATP-grasp target RiPP | | Authors: | Link, A.J, Choi, B. | | Deposit date: | 2022-08-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mechanistic Analysis of the Biosynthesis of the Aspartimidylated Graspetide Amycolimiditide.
J.Am.Chem.Soc., 144, 2022
|
|
7KS6
 
 | | STRUCTURE OF TETRASACCHARIDE BUILDING BLOCK OF A SULFATED FUCAN FROM LYTECHINUS VARIEGATUS | | Descriptor: | 4-O-sulfo-alpha-L-fucopyranose-(1-3)-2,4-di-O-sulfo-alpha-L-fucopyranose-(1-3)-2-O-sulfo-alpha-L-fucopyranose-(1-3)-2-O-sulfo-alpha-L-fucopyranose | | Authors: | Kim, S.B, Thara, R, Aderibigbe, A.O, Doerksen, R.J, Pomin, V.H. | | Deposit date: | 2020-11-21 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformational properties of l-fucose and the tetrasaccharide building block of the sulfated l-fucan from Lytechinus variegatus.
J.Struct.Biol., 209, 2020
|
|
6ZGG
 
 | |
3JA7
 
 | | Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution | | Descriptor: | Portal protein gp20 | | Authors: | Sun, L, Zhang, X, Gao, S, Rao, P.A, Padilla-Sanchez, V, Chen, Z, Sun, S, Xiang, Y, Subramaniam, S, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2015-04-21 | | Release date: | 2015-07-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution.
Nat Commun, 6, 2015
|
|
3K7Y
 
 | | Aspartate Aminotransferase of Plasmodium falciparum | | Descriptor: | ACETATE ION, Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Groves, M.R, Jordanova, R, Jain, R, Wrenger, C, Muller, I.B. | | Deposit date: | 2009-10-13 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Specific Inhibition of the Aspartate Aminotransferase of Plasmodium falciparum.
J.Mol.Biol., 405, 2011
|
|
4V7G
 
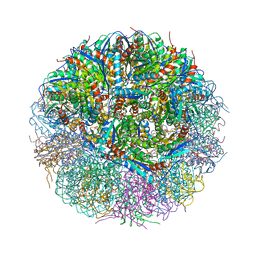 | | Crystal Structure of Lumazine Synthase from Bacillus Anthracis | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Morgunova, E, Illarionov, B, Saller, S, Popov, A, Sambaiah, T, Bacher, A, Cushman, M, Fischer, M, Ladenstein, R. | | Deposit date: | 2009-09-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural study and thermodynamic characterization of inhibitor binding to lumazine synthase from Bacillus anthracis.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3K73
 
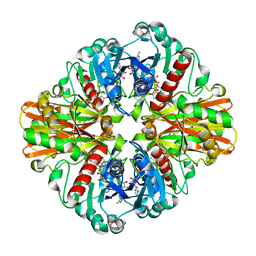 | | Crystal Structure of Phosphate bound Holo Glyceraldehyde-3-phosphate dehydrogenase 1 from MRSA252 at 2.5 Angstrom resolution | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2009-10-12 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
6TR2
 
 | |
6Z1X
 
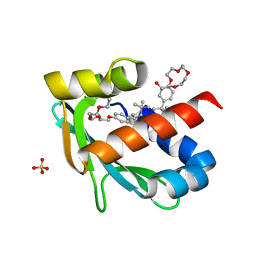 | |
6Z1W
 
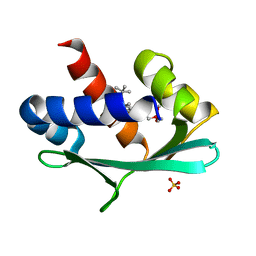 | |
5OGA
 
 | | Structure of minimal i-motif domain | | Descriptor: | DNA (5'-D(*TP*(DCP)P*GP*TP*TP*CP*(DCP)P*GP*TP*TP*TP*TP*TP*CP*GP*TP*TP*CP*CP*GP*T)-3') | | Authors: | Mir, B, Serrano, I, Buitrago, D, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2017-07-12 | | Release date: | 2017-11-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Prevalent Sequences in the Human Genome Can Form Mini i-Motif Structures at Physiological pH.
J. Am. Chem. Soc., 139, 2017
|
|
5OBN
 
 | | NMR solution structure of U11/U12 65K protein's C-terminal RRM domain (381-516) | | Descriptor: | RNA-binding protein 40 | | Authors: | Norppa, A.J, Kauppala, T.M, Heikkinen, H.A, Verma, B, Iwai, H, Frilander, M.J. | | Deposit date: | 2017-06-28 | | Release date: | 2018-01-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Mutations in the U11/U12-65K protein associated with isolated growth hormone deficiency lead to structural destabilization and impaired binding of U12 snRNA.
RNA, 24, 2018
|
|
6Z4N
 
 | | CRYSTAL STRUCTURE OF OASS COMPLEXED WITH UPAR INHIBITOR | | Descriptor: | (1~{S},2~{S})-1-[(4-methylphenyl)methyl]-2-phenyl-cyclopropane-1-carboxylic acid, COBALT (II) ION, Cysteine synthase A, ... | | Authors: | Demitri, N, Storici, P, Campanini, B. | | Deposit date: | 2020-05-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Investigational Studies on a Hit Compound Cyclopropane-Carboxylic Acid Derivative Targeting O -Acetylserine Sulfhydrylase as a Colistin Adjuvant.
Acs Infect Dis., 7, 2021
|
|
6TH8
 
 | |
6TPB
 
 | | NMR structure of the apo-form of Pseudomonas fluorescens CopC | | Descriptor: | Putative copper resistance protein | | Authors: | Persson, K.C, Mayzel, M, Karlsson, B.G, Peciulyte, A, Olsson, L, Wittung Stafshede, P, Salomon Johansen, K, Horvath, I. | | Deposit date: | 2019-12-13 | | Release date: | 2021-01-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Pseudomonas fluorescens CopC
To Be Published
|
|
4V4Z
 
 | | 70S Thermus thermophilous ribosome functional complex with mRNA and E- and P-site tRNAs at 4.5A. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Jenner, L, Yusupova, G, Rees, B, Moras, D, Yusupov, M. | | Deposit date: | 2006-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.51 Å) | | Cite: | Structural basis for messenger RNA movement on the ribosome.
Nature, 444, 2006
|
|
4V5T
 
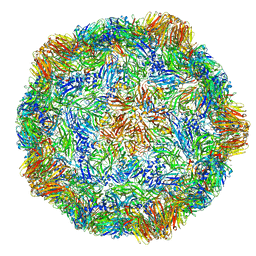 | | X-ray structure of the Grapevine Fanleaf virus | | Descriptor: | COAT PROTEIN | | Authors: | Schellenberger, P, Sauter, C, Lorber, B, Bron, P, Trapani, S, Bergdoll, M, Marmonier, A, Schmitt-Keichinger, C, Lemaire, O, Demangeat, G, Ritzenthaler, C. | | Deposit date: | 2011-02-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights Into Viral Determinants of Nematode Mediated Grapevine Fanleaf Virus Transmission.
Plos Pathog., 7, 2011
|
|
3KFO
 
 | | Crystal structure of the C-terminal domain from the nuclear pore complex component NUP133 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Nucleoporin NUP133 | | Authors: | Sampathkumar, P, Bonanno, J.B, Miller, S, Bain, K, Dickey, M, Gheyi, T, Almo, S.C, Rout, M, Sali, A, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-27 | | Release date: | 2010-01-26 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the C-terminal domain of Saccharomyces cerevisiae Nup133, a component of the nuclear pore complex.
Proteins, 79, 2011
|
|
6ZGN
 
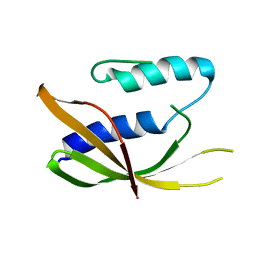 | | Crystal structure of VirB8-like OrfG central domain of Streptococcus thermophilus ICESt3; a putative assembly factor of a gram positive conjugative Type IV secretion system. | | Descriptor: | Putative transfer protein | | Authors: | Cappele, J, Mohamad-Ali, A, Leblond-Bourget, N, Payot-Lacroix, S, Mathiot, S, Didierjean, C, Favier, F, Douzi, B. | | Deposit date: | 2020-06-19 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Biochemical Analysis of OrfG: The VirB8-like Component of the Conjugative Type IV Secretion System of ICE St3 From Streptococcus thermophilus .
Front Mol Biosci, 8, 2021
|
|
3KDS
 
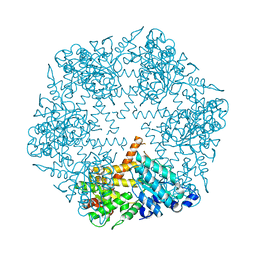 | | apo-FtsH crystal structure | | Descriptor: | Cell division protein FtsH, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-naphthalen-2-yl-L-alanyl-L-alaninamide, ZINC ION | | Authors: | Bieniossek, C, Niederhauser, B, Baumann, U. | | Deposit date: | 2009-10-23 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | The crystal structure of apo-FtsH reveals domain movements necessary for substrate unfolding and translocation
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5OOU
 
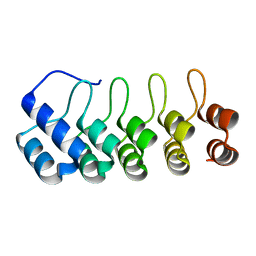 | | Designed Ankyrin Repeat Protein (DARPin) YTRL-1 selected by directed evolution against Lysozyme | | Descriptor: | DARPin YTRL-1 | | Authors: | Fischer, G, Hogan, B.J, Houlihan, G, Edmond, S, Huovinen, T.T.K, Hollfelder, F, Hyvonen, M. | | Deposit date: | 2017-08-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Designed Ankyrin Repeat Protein (DARPin) YTRL-1 selected by directed evolution against Lysozyme
To be published
|
|
3KBE
 
 | | Metal-free C. elegans Cu,Zn Superoxide Dismutase | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Pakhomova, O.N, Taylor, A.B, Schuermann, J.P, Culotta, V.L, Hart, P.J. | | Deposit date: | 2009-10-20 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray Crystal Structure of C. elegans Cu,Zn Superoxide Dismutase
To be Published
|
|
3KJT
 
 | |
8J9G
 
 | | CrtSPARTA hetero-dimer bound with guide-target, state 1 | | Descriptor: | DNA (25-MER), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Li, Z.X, Guo, L.J, Huang, P.P, Xiao, Y.B, Chen, M.R. | | Deposit date: | 2023-05-03 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Auto-inhibition and activation of a short Argonaute-associated TIR-APAZ defense system.
Nat.Chem.Biol., 20, 2024
|
|
3KLK
 
 | | Crystal structure of Lactobacillus reuteri N-terminally truncated glucansucrase GTF180 in triclinic apo- form | | Descriptor: | CALCIUM ION, GLYCEROL, Glucansucrase | | Authors: | Vujicic-Zagar, A, Pijning, T, Kralj, S, Eeuwema, W, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2009-11-08 | | Release date: | 2010-11-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
