2LF6
 
 | | Solution NMR structure of HopABPph1448_220_320 from Pseudomonas syringae pv. phaseolicola str. 1448A, Midwest Center for Structural Genomics target APC40132.4 and Northeast Structural Genomics Consortium target PsT3A | | Descriptor: | Effector protein hopAB1 | | Authors: | Wu, B, Yee, A, Houliston, S, Semesi, A, Garcia, M, Singer, A.U, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
2M46
 
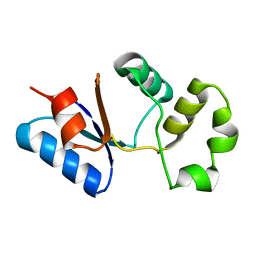 | | Solution NMR structure of SACOL0876 from Staphylococcus aureus COL, NESG target ZR353 and CSGID target IDP00841 | | Descriptor: | Arsenate reductase, putative | | Authors: | Wu, B, Yee, A, Houliston, S, Garcia, M, Savchenko, A, Arrowsmith, C.H, Anderson, W.F, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of SACOL0876 from Staphylococcus aureus COL, NESG target ZR353 and CSGID target IDP00841
To be Published
|
|
6Y4U
 
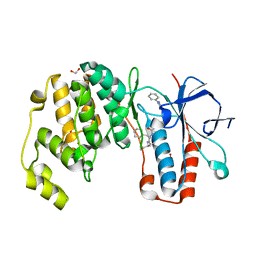 | | Crystal structure of p38 in complex with SR65 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-oxidanylidene-1-(pentan-3-ylamino)butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Roehm, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
6Y2D
 
 | | Crystal structure of the second KH domain of FUBP1 | | Descriptor: | Far upstream element-binding protein 1, GLYCEROL, SULFATE ION | | Authors: | Ni, X, Chaikuad, A, Joerger, A.C, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparative structural analyses and nucleotide-binding characterization of the four KH domains of FUBP1.
Sci Rep, 10, 2020
|
|
6Y5V
 
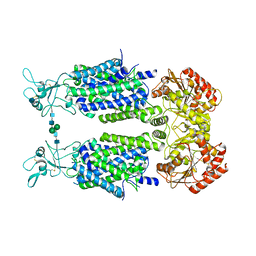 | | Structure of Human Potassium Chloride Transporter KCC3b (S45D/T940D/T997D) in KCl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier family 12 member 6, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chi, G, Man, H, Ebenhoch, R, Reggiano, G, Pike, A.C.W, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, E.M, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Singh, N.K, Abrusci, P, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, DiMaio, F, Duerr, K.L. | | Deposit date: | 2020-02-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Phospho-regulation, nucleotide binding and ion access control in potassium-chloride cotransporters.
Embo J., 40, 2021
|
|
2M4E
 
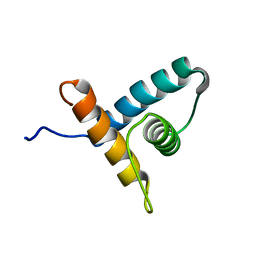 | | Solution NMR structure of VV2_0175 from Vibrio vulnificus, NESG target VnR1 and CSGID target IDP91333 | | Descriptor: | Putative uncharacterized protein | | Authors: | Wu, B, Yee, A, Houliston, S, Lemak, A, Garcia, M, Savchenko, A, Arrowsmith, C.H, Anderson, W.F, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-04 | | Release date: | 2013-03-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of VV2_0175 from Vibrio vulnificus, NESG target VnR1 and CSGID target IDP91333
To be Published
|
|
6Y2C
 
 | | Crystal structure of the third KH domain of FUBP1 | | Descriptor: | 1,2-ETHANEDIOL, Far upstream element-binding protein 1, ZINC ION | | Authors: | Ni, X, Joerger, A.C, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparative structural analyses and nucleotide-binding characterization of the four KH domains of FUBP1.
Sci Rep, 10, 2020
|
|
6Y4T
 
 | | Crystal structure of p38 in complex with SR63. | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-1-[[(2~{S})-butan-2-yl]amino]-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Roehm, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
6Y4V
 
 | | Crystal structure of p38 in complex with SR68 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-(3-methylbutylamino)-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Roehm, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
6Y3U
 
 | | Crystal structure of PPARgamma in complex with compound (R)-16 | | Descriptor: | (2~{R})-2-[[6-[(2,4-dichlorophenyl)sulfonylamino]-1,3-benzothiazol-2-yl]sulfanyl]octanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Hanke, T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-18 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | A Selective Modulator of Peroxisome Proliferator-Activated Receptor gamma with an Unprecedented Binding Mode.
J.Med.Chem., 63, 2020
|
|
6Y6V
 
 | | p38a bound with MCP-81 | | Descriptor: | 5-azanyl-~{N}-[[4-[[5-~{tert}-butyl-2-(4-methylphenyl)pyrazol-3-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Schroeder, M, Roehm, S, Knapp, S, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-27 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
6Y24
 
 | | Crystal structure of fourth KH domain of FUBP1 | | Descriptor: | 1,2-ETHANEDIOL, Far upstream element-binding protein 1 | | Authors: | Ni, X, Joerger, A.C, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Comparative structural analyses and nucleotide-binding characterization of the four KH domains of FUBP1.
Sci Rep, 10, 2020
|
|
6Y6H
 
 | | Crystal structure of STK17b (DRAK2) in complex with UNC-AP-194 probe | | Descriptor: | 1,2-ETHANEDIOL, 2-[6-(1-benzothiophen-2-yl)thieno[3,2-d]pyrimidin-4-yl]sulfanylethanoic acid, Serine/threonine-protein kinase 17B | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Drewry, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Chemical Probe for Dark Kinase STK17B Derives Its Potency and High Selectivity through a Unique P-Loop Conformation.
J.Med.Chem., 63, 2020
|
|
6Y4W
 
 | | Crystal structure of p38 in complex with SR69 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-(cyclohexylamino)-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Roehm, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
6Y6F
 
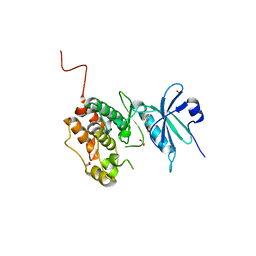 | | Crystal structure of STK17B (DRAK2) in complex with PKIS43 | | Descriptor: | 1,2-ETHANEDIOL, 2-[6-(4-methylsulfanylphenyl)thieno[3,2-d]pyrimidin-4-yl]sulfanylethanoic acid, Serine/threonine-protein kinase 17B | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Drewry, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A Chemical Probe for Dark Kinase STK17B Derives Its Potency and High Selectivity through a Unique P-Loop Conformation.
J.Med.Chem., 63, 2020
|
|
6YHR
 
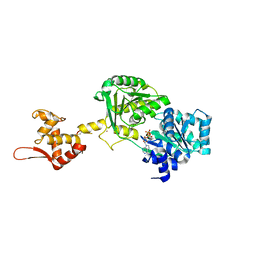 | | Crystal structure of Werner syndrome helicase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Werner syndrome ATP-dependent helicase, ZINC ION | | Authors: | Newman, J.A, Gavard, A.E, Savitsky, P, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-03-30 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the helicase core of Werner helicase, a key target in microsatellite instability cancers.
Life Sci Alliance, 4, 2021
|
|
6YMQ
 
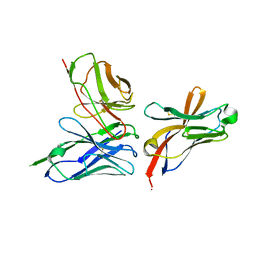 | | TREM2 extracellular domain (19-131) in complex with single-chain variable 4 (scFv-4) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Single-chain variable 4, ... | | Authors: | Szykowska, A, Preger, C, Scacioc, A, Mukhopadhyay, S.M.M, McKinley, G, Graslund, S, Wigren, E, Persson, H, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Di Daniel, E, Davis, J.B, Burgess-Brown, N, Bullock, A. | | Deposit date: | 2020-04-09 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Selection and structural characterization of anti-TREM2 scFvs that reduce levels of shed ectodomain.
Structure, 29, 2021
|
|
6Y6C
 
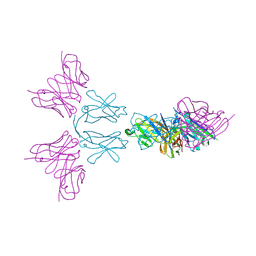 | | TREM2 extracellular domain (19-174) in complex with single-chain variable fragment (scFv-4) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Single chain variable, Triggering receptor expressed on myeloid cells 2 | | Authors: | Szykowska, A, Preger, C, Williams, E, Mukhopadhyay, S.M.M, McKinley, G, Gruslund, S, Wigren, E, Persson, H, Arrowsmith, C.H, Edwards, A, von Delft, F, Bountra, C, Davis, J.B, Di Daniel, E, Burgess-Brown, N, Bullock, A. | | Deposit date: | 2020-02-26 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Selection and structural characterization of anti-TREM2 scFvs that reduce levels of shed ectodomain.
Structure, 29, 2021
|
|
6XKB
 
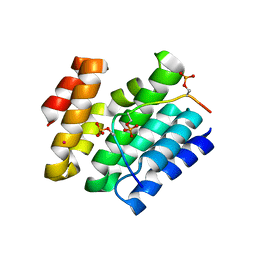 | | Crystal structure of SR-related and CTD-associated factor 4(SCAF4-CID)with peptide S2,S5p-CTD | | Descriptor: | S2,S5p-CTD peptide, SR-related and CTD-associated factor 4, UNKNOWN ATOM OR ION | | Authors: | Zhou, M.Q, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-26 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of the S2, S5-phosphorylated RNA polymerase II CTD by the mRNA anti-terminator protein hSCAF4.
Febs Lett., 596, 2022
|
|
6XY7
 
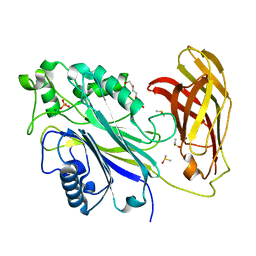 | | Human SHIP1 with magnesium and phosphate bound to the active site | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Bradshaw, W.J, Scacioc, A, Fernandez-Cid, A, Mckinley, G, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-01-29 | | Release date: | 2020-02-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.086 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
6Y4X
 
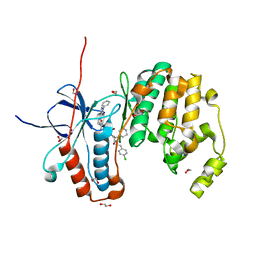 | | Crystal structure of p38 in complex with SR72 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-1-[2-(4-chlorophenyl)ethylamino]-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Roehm, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
6ZEL
 
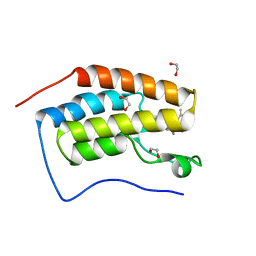 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound F5 | | Descriptor: | 1,2-ETHANEDIOL, 3,5-dimethyl-4-[(6-methylpyrimidin-4-yl)sulfanylmethyl]-1,2-oxazole, Bromodomain-containing protein 4 | | Authors: | Krojer, T, Martinez-Cartro, M, Picaud, S, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Barril, X, von Delft, F. | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound F5
To Be Published
|
|
6XKC
 
 | | Crystal structure of E3 ligase | | Descriptor: | Protein fem-1 homolog C | | Authors: | Yan, X, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Dong, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular basis for ubiquitin ligase CRL2 FEM1C -mediated recognition of C-degron.
Nat.Chem.Biol., 17, 2021
|
|
6ZED
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound F1 | | Descriptor: | 1,2-ETHANEDIOL, 4,5-dimethyl-1~{H}-pyrazolo[3,4-c]pyridazin-3-amine, Bromodomain-containing protein 4 | | Authors: | Krojer, T, Martinez-Cartro, M, Picaud, S, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Barril, X, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound F1
To Be Published
|
|
6ZF9
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound SSR4 | | Descriptor: | 1,2-ETHANEDIOL, 3-ethyl-5-phenyl-[1,3]thiazolo[2,3-c][1,2,4]triazole, Bromodomain-containing protein 4 | | Authors: | Krojer, T, Martinez-Cartro, M, Picaud, S, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Barril, X, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound SSR4
To Be Published
|
|
