1WDA
 
 | | Crystal structure of human peptidylarginine deiminase type4 (PAD4) in complex with benzoyl-L-arginine amide | | Descriptor: | CALCIUM ION, N-[(E)-2-AMINO-1-(3-{[AMINO(IMINO)METHYL]AMINO}PROPYL)-2-HYDROXYVINYL]BENZAMIDE, Protein-arginine deiminase type IV, ... | | Authors: | Arita, K, Hashimoto, H, Shimizu, T, Nakashima, K, Yamada, M, Sato, M. | | Deposit date: | 2004-05-12 | | Release date: | 2004-07-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for Ca(2+)-induced activation of human PAD4
Nat.Struct.Mol.Biol., 11, 2004
|
|
1WD9
 
 | | Calcium bound form of human peptidylarginine deiminase type4 (PAD4) | | Descriptor: | CALCIUM ION, Protein-arginine deiminase type IV, SULFATE ION | | Authors: | Arita, K, Hashimoto, H, Shimizu, T, Nakashima, K, Yamada, M, Sato, M. | | Deposit date: | 2004-05-12 | | Release date: | 2004-07-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for Ca(2+)-induced activation of human PAD4
Nat.Struct.Mol.Biol., 11, 2004
|
|
1WD8
 
 | | Calcium free form of human peptidylarginine deiminase type4 (PAD4) | | Descriptor: | Protein-arginine deiminase type IV | | Authors: | Arita, K, Hashimoto, H, Shimizu, T, Nakashima, K, Yamada, M, Sato, M. | | Deposit date: | 2004-05-12 | | Release date: | 2004-07-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for Ca(2+)-induced activation of human PAD4
Nat.Struct.Mol.Biol., 11, 2004
|
|
6IIW
 
 | | Crystal structure of human UHRF1 PHD finger in complex with PAF15 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, E3 ubiquitin-protein ligase UHRF1, PCNA-associated factor, ... | | Authors: | Arita, K, Kori, S. | | Deposit date: | 2018-10-07 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Two distinct modes of DNMT1 recruitment ensure stable maintenance DNA methylation.
Nat Commun, 11, 2020
|
|
2E1N
 
 | |
2DEX
 
 | | Crystal structure of human peptidylarginine deiminase 4 in complex with histone H3 N-terminal peptide including Arg17 | | Descriptor: | 10-mer peptide from histone H3, CALCIUM ION, Protein-arginine deiminase type IV, ... | | Authors: | Arita, K, Shimizu, T, Hashimoto, H, Hidaka, Y, Yamada, M, Sato, M. | | Deposit date: | 2006-02-18 | | Release date: | 2006-04-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for histone N-terminal recognition by human peptidylarginine deiminase 4
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2DEW
 
 | | Crystal structure of human peptidylarginine deiminase 4 in complex with histone H3 N-terminal tail including Arg8 | | Descriptor: | 10-mer peptide from histone H3, CALCIUM ION, Protein-arginine deiminase type IV, ... | | Authors: | Arita, K, Shimizu, T, Hashimoto, H, Hidaka, Y, Yamada, M, Sato, M. | | Deposit date: | 2006-02-18 | | Release date: | 2006-04-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for histone N-terminal recognition by human peptidylarginine deiminase 4
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2DEY
 
 | | Crystal structure of human peptidylarginine deiminase 4 in complex with histone H4 N-terminal tail including Arg3 | | Descriptor: | 10-mer peptide from histone H4, CALCIUM ION, Protein-arginine deiminase type IV, ... | | Authors: | Arita, K, Shimizu, T, Hashimoto, H, Hidaka, Y, Yamada, M, Sato, M. | | Deposit date: | 2006-02-18 | | Release date: | 2006-04-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for histone N-terminal recognition by human peptidylarginine deiminase 4
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3ASK
 
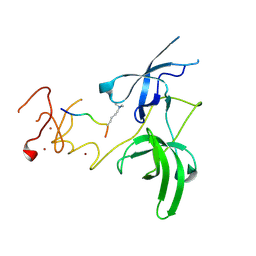 | | Structure of UHRF1 in complex with histone tail | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Histone H3.3, ZINC ION | | Authors: | Arita, K, Sugita, K, Unoki, M, Hamamoto, R, Sekiyama, N, Tochio, H, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2010-12-16 | | Release date: | 2012-01-25 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ASL
 
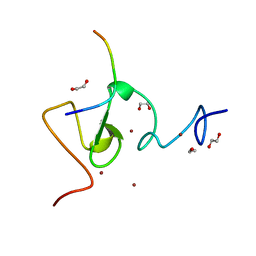 | | Structure of UHRF1 in complex with histone tail | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1, Histone H3.3, ... | | Authors: | Arita, K, Sugita, K, Unoki, M, Hamamoto, R, Sekiyama, N, Tochio, H, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2010-12-16 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2ZKE
 
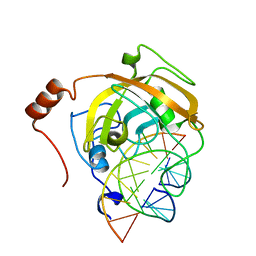 | | Crystal structure of the SRA domain of mouse Np95 in complex with hemi-methylated CpG DNA | | Descriptor: | DNA (5'-D(*DCP*DTP*DAP*DCP*DCP*DGP*DGP*DAP*DTP*DTP*DGP*DC)-3'), DNA (5'-D(*DGP*DCP*DAP*DAP*DTP*DCP*(5CM)P*DGP*DGP*DTP*DAP*DG)-3'), E3 ubiquitin-protein ligase UHRF1 | | Authors: | Arita, K, Ariyoshi, M, Tochio, H, Nakamura, Y, Shirakawa, M. | | Deposit date: | 2008-03-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism
Nature, 455, 2008
|
|
2ZKF
 
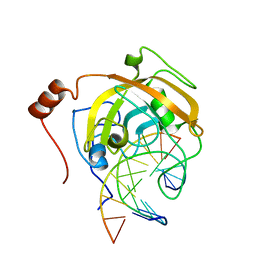 | | Crystal structure of the SRA domain of mouse Np95 in complex with hemi-methylated CpG DNA | | Descriptor: | DNA (5'-D(*DCP*DTP*DAP*DTP*DCP*(5CM)P*DGP*DGP*DTP*DGP*DA)-3'), DNA (5'-D(P*DCP*DAP*DCP*DCP*DGP*DGP*DAP*DTP*DAP*DGP*DA)-3'), E3 ubiquitin-protein ligase UHRF1 | | Authors: | Arita, K, Ariyoshi, M, Tochio, H, Nakamura, Y, Shirakawa, M. | | Deposit date: | 2008-03-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism
Nature, 455, 2008
|
|
2ZKD
 
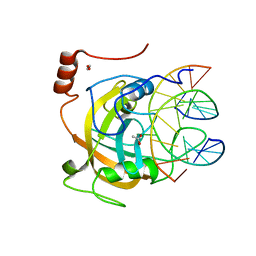 | | Crystal structure of the SRA domain of mouse Np95 in complex with hemi-methylated CpG DNA | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*DCP*DTP*DAP*DCP*DCP*DGP*DGP*DAP*DTP*DTP*DGP*DC)-3'), ... | | Authors: | Arita, K, Ariyoshi, M, Tochio, H, Nakamura, Y, Shirakawa, M. | | Deposit date: | 2008-03-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism
Nature, 455, 2008
|
|
2ZKG
 
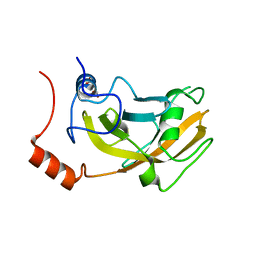 | | Crystal structure of unliganded SRA domain of mouse Np95 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1 | | Authors: | Arita, K, Ariyoshi, M, Tochio, H, Nakamura, Y, Shirakawa, M. | | Deposit date: | 2008-03-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism
Nature, 455, 2008
|
|
2CVD
 
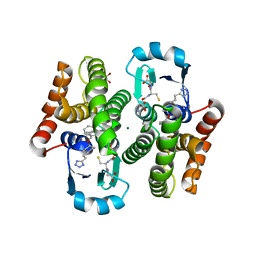 | | Crystal structure analysis of human hematopoietic prostaglandin D synthase complexed with HQL-79 | | Descriptor: | 4-(BENZHYDRYLOXY)-1-[3-(1H-TETRAAZOL-5-YL)PROPYL]PIPERIDINE, GLUTATHIONE, GLYCEROL, ... | | Authors: | Aritake, K, Kado, Y, Inoue, T, Miyano, M, Urade, Y. | | Deposit date: | 2005-06-02 | | Release date: | 2006-04-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Functional Characterization of HQL-79, an Orally Selective Inhibitor of Human Hematopoietic Prostaglandin D Synthase.
J.Biol.Chem., 281, 2006
|
|
8YV8
 
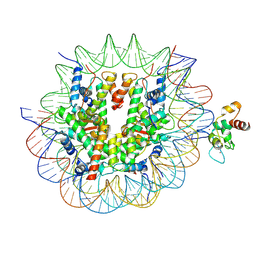 | | Cryo-EM structure of CDCA7 bound to nucleosome including hemimethylated CpG site in Widom601 positioning sequence. | | Descriptor: | Cell division cycle-associated protein 7, DNA (132-MER), Histone H2A type 1-B/E, ... | | Authors: | Kikuchi, A, Shikimachi, R, Nishiyama, A, Funabiki, H, Arita, K. | | Deposit date: | 2024-03-28 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | CDCA7 is a hemimethylated DNA adaptor for the nucleosome remodeler HELLS
To Be Published
|
|
5WVO
 
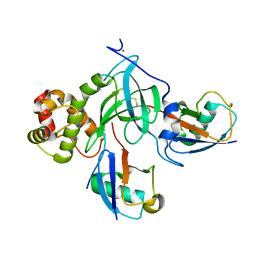 | | Crystal structure of DNMT1 RFTS domain in complex with K18/K23 mono-ubiquitylated histone H3 | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, Histone H3.1, Ubiquitin, ... | | Authors: | Ishiyama, S, Nishiyama, A, Nakanishi, M, Arita, K. | | Deposit date: | 2016-12-28 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structure of the Dnmt1 Reader Module Complexed with a Unique Two-Mono-Ubiquitin Mark on Histone H3 Reveals the Basis for DNA Methylation Maintenance
Mol. Cell, 68, 2017
|
|
7XGA
 
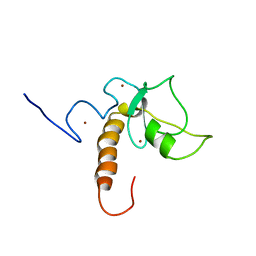 | |
7XI9
 
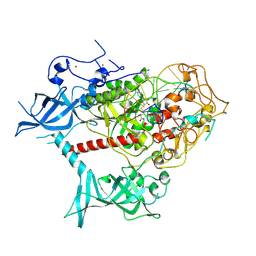 | | Cryo-EM structure of human DNMT1 (aa:351-1616) in complex with ubiquitinated H3 and hemimethylated DNA analog (CXXC-ordered form) | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*AP*(5CM)P*GP*GP*AP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*TP*CP*(C55)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Onoda, H, Kikuchi, A, Kori, S, Yoshimi, S, Yamagata, A, Arita, K. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural basis for activation of DNMT1.
Nat Commun, 13, 2022
|
|
7XIB
 
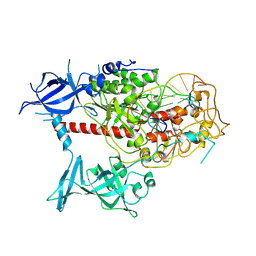 | | Cryo-EM structure of human DNMT1 (aa:351-1616) in complex with ubiquitinated H3 and hemimethylated DNA analog (CXXC-disordered form) | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*AP*(5CM)P*GP*GP*AP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*TP*CP*(C55)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Onoda, H, Kikuchi, A, Kori, S, Yoshimi, S, Yamagata, A, Arita, K. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | Structural basis for activation of DNMT1.
Nat Commun, 13, 2022
|
|
8WMS
 
 | |
2RRU
 
 | | Solution structure of the UBA omain of p62 and its interaction with ubiquitin | | Descriptor: | Sequestosome-1 | | Authors: | Isogai, S, Morimoto, D, Arita, K, Unzai, S, Tenno, T, Hasegawa, J, Sou, Y, Komatsu, M, Tanaka, K, Shirakawa, M, Tochio, H. | | Deposit date: | 2011-06-09 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal structure of the UBA omain of p62 and its interaction with ubiquitin
To be Published
|
|
7FB7
 
 | | Crystal structure of human UHRF1 TTD in complex with 5-amino-2,4-dimethylpyridine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-amino-2,4-dimethylpyridine, DIMETHYL SULFOXIDE, ... | | Authors: | Kori, S, Arita, K, Yoshimi, S. | | Deposit date: | 2021-07-08 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-based screening combined with computational and biochemical analyses identified the inhibitor targeting the binding of DNA Ligase 1 to UHRF1.
Bioorg.Med.Chem., 52, 2021
|
|
5YY9
 
 | | Crystal structure of Tandem Tudor Domain of human UHRF1 in complex with LIG1-K126me3 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Ligase 1 | | Authors: | Kori, S, Defossez, P.A, Arita, K. | | Deposit date: | 2017-12-08 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Structure of the UHRF1 Tandem Tudor Domain Bound to a Methylated Non-histone Protein, LIG1, Reveals Rules for Binding and Regulation.
Structure, 27, 2019
|
|
5YYA
 
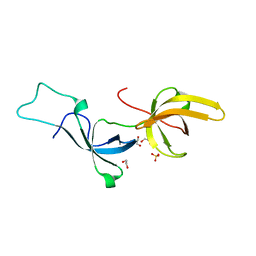 | | Crystal structure of Tandem Tudor Domain of human UHRF1 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1, SULFATE ION | | Authors: | Kori, S, Defossez, P.A, Arita, K. | | Deposit date: | 2017-12-08 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the UHRF1 Tandem Tudor Domain Bound to a Methylated Non-histone Protein, LIG1, Reveals Rules for Binding and Regulation.
Structure, 27, 2019
|
|
