6OMG
 
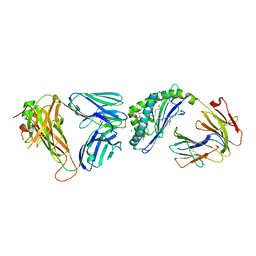 | | Structure of mouse CD1D- Glc-DAG (sn-1 C18:0, sn-2 C18:1c9)-iNKT TCR Ternary complex | | Descriptor: | (2R)-1-(alpha-D-glucopyranosyloxy)-3-(octadecanoyloxy)propan-2-yl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dirk, M.Z, Bitra, A, Wang, J. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of mouse CD1D- Glc-DAG (sn-1 C18:0, sn-2 C18:1c9)-iNKT TCR Ternary complex
To Be Published
|
|
4U27
 
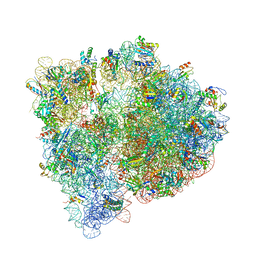 | | Crystal structure of the E. coli ribosome bound to flopristin and linopristin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Noeske, J, Huang, J, Olivier, N.B, Giacobbe, R.A, Zambrowski, M, Cate, J.H.D. | | Deposit date: | 2014-07-16 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Synergy of streptogramin antibiotics occurs independently of their effects on translation.
Antimicrob.Agents Chemother., 58, 2014
|
|
8D4K
 
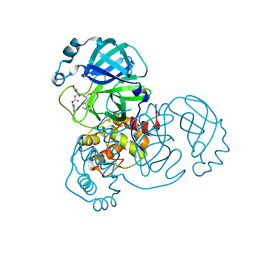 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H172Y Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4M
 
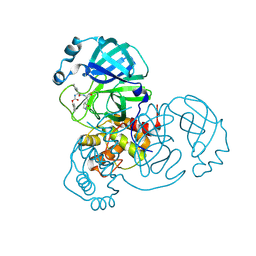 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144A Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
1YEM
 
 | | Conserved hypothetical protein Pfu-838710-001 from Pyrococcus furiosus | | Descriptor: | Conserved hypothetical protein Pfu-838710-001, PLATINUM (II) ION, UNKNOWN ATOM OR ION | | Authors: | Yang, H, Chang, J, Shah, A, Ng, J.D, Liu, Z.-J, Chen, L, Lee, D, Tempel, W, Praissman, J.L, Lin, D, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-28 | | Release date: | 2005-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conserved hypothetical protein Pfu-838710-001 from Pyrococcus furiosus
To be published
|
|
8D4J
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H172Y Mutant | | Descriptor: | 3C-like proteinase nsp5, GLYCEROL | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4N
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) E166Q Mutant | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
4U1U
 
 | | Crystal structure of the E. coli ribosome bound to quinupristin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Noeske, J, Huang, J, Olivier, N.B, Giacobbe, R.A, Zambrowski, M, Cate, J.H.D. | | Deposit date: | 2014-07-16 | | Release date: | 2014-07-30 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Synergy of streptogramin antibiotics occurs independently of their effects on translation.
Antimicrob.Agents Chemother., 58, 2014
|
|
8D8O
 
 | | Cryo-EM structure of substrate unbound PAPP-A | | Descriptor: | Pappalysin-1, ZINC ION | | Authors: | Judge, R.A, Jain, R, Hao, Q, Ouch, C, Sridar, J, Smith, C.L, Wang, J.C.K, Eaton, D. | | Deposit date: | 2022-06-08 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure of the PAPP-ABP5 complex reveals mechanism of substrate recognition
Nat Commun, 13, 2022
|
|
4U1V
 
 | | Crystal structure of the E. coli ribosome bound to linopristin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Noeske, J, Huang, J, Olivier, N.B, Giacobbe, R.A, Zambrowski, M, Cate, J.H.D. | | Deposit date: | 2014-07-16 | | Release date: | 2014-07-30 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synergy of streptogramin antibiotics occurs independently of their effects on translation.
Antimicrob.Agents Chemother., 58, 2014
|
|
4U7T
 
 | | Crystal structure of DNMT3A-DNMT3L in complex with histone H3 | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Guo, X, Wang, L, Yin, X, Li, J, Xiao, J, He, S, Wang, J, Xu, Y. | | Deposit date: | 2014-07-31 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insight into autoinhibition and histone H3-induced activation of DNMT3A
Nature, 517, 2015
|
|
5N8C
 
 | | Crystal structure of Pseudomonas aeruginosa LpxC complexed with inhibitor | | Descriptor: | (2~{S})-3-azanyl-2-[[(1~{R})-5-[2-[4-[[2-(hydroxymethyl)imidazol-1-yl]methyl]phenyl]ethynyl]-2,3-dihydro-1~{H}-inden-1-yl]amino]-3-methyl-~{N}-oxidanyl-butanamide, CHLORIDE ION, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Cross, J.B, Ryan, M.D, Zhang, J, Cheng, R.K, Wood, M, Andersen, O.A, Brooks, M, Kwong, J, Barker, J. | | Deposit date: | 2017-02-23 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based discovery of LpxC inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
1YB3
 
 | | Conserved hypothetical protein Pfu-178653-001 from Pyrococcus furiosus | | Descriptor: | UNKNOWN ATOM OR ION, hypothetical protein | | Authors: | Habel, J, Zhou, W, Chang, J, Zhao, M, Xu, H, Chen, L, Lee, D, Nguyen, D, Chang, S.-H, Horanyi, P, Florence, Q, Tempel, W, Lin, D, Zhang, H, Praissman, J, Jenney Jr, F.E, Adams, M.W.W, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-19 | | Release date: | 2005-02-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conserved hypothetical protein Pfu-178653-001 from Pyrococcus furiosus
To be published
|
|
1YBX
 
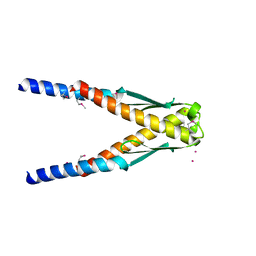 | | Conserved hypothetical protein Cth-383 from Clostridium thermocellum | | Descriptor: | Conserved hypothetical protein, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Chang, J, Zhao, M, Habel, J, Kataeva, I, Xu, H, Chen, L, Lee, D, Nguyen, J, Chang, S.-H, Horanyi, P, Florence, Q, Zhou, W, Lin, D, Zhang, H, Ljundahl, L, Liu, Z.-J, Rose, J, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conserved hypothetical protein Cth-383 from Clostridium thermocellum
To be published
|
|
1YD7
 
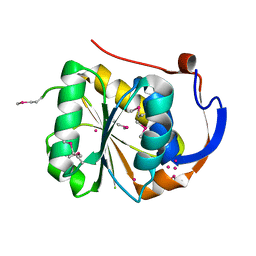 | | Conserved hypothetical protein Pfu-1647980-001 from Pyrococcus furiosus | | Descriptor: | 2-keto acid:ferredoxin oxidoreductase subunit alpha, UNKNOWN ATOM OR ION | | Authors: | Horanyi, P, Florence, Q, Zhou, W, Chang, J, Zhao, M, Habel, J, Xu, H, Chen, L, Lee, D, Nguyen, D, Chang, S.-H, Tempel, W, Lin, D, Zhang, H, Praissman, J, Jenney Jr, F.E, Adams, M.W.W, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-23 | | Release date: | 2005-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conserved hypothetical protein Pfu-1647980-001 from Pyrococcus furiosus
To be published
|
|
4U7U
 
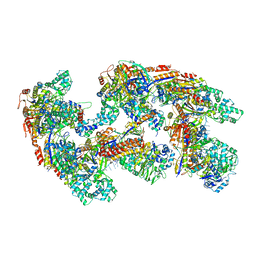 | | Crystal structure of RNA-guided immune Cascade complex from E.coli | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Zhao, H, Sheng, G, Wang, J, Wang, M, Bunkoczi, G, Gong, W, Wei, Z, Wang, Y. | | Deposit date: | 2014-07-31 | | Release date: | 2014-08-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli
Nature, 515, 2014
|
|
1ZGH
 
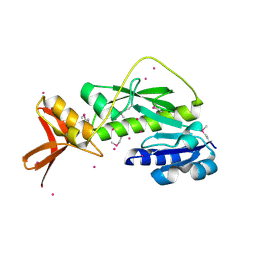 | | Methionyl-tRNA formyltransferase from Clostridium thermocellum | | Descriptor: | Methionyl-tRNA formyltransferase, UNKNOWN ATOM OR ION | | Authors: | Yang, H, Kataeva, I, Xu, H, Zhao, M, Chang, J, Liu, Z, Chen, L, Tempel, W, Habel, J, Zhou, W, Lee, D, Lin, D, Chang, S, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Wang, B, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-04-21 | | Release date: | 2005-05-03 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Methionyl-tRNA formyltransferase from Clostridium thermocellum
To be published
|
|
7TOB
 
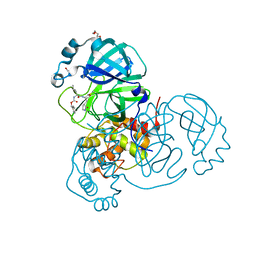 | | Crystal structure of the SARS-CoV-2 Omicron main protease (Mpro) in complex with inhibitor GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Sacco, M.D, Wang, J, Chen, Y. | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The P132H mutation in the main protease of Omicron SARS-CoV-2 decreases thermal stability without compromising catalysis or small-molecule drug inhibition.
Cell Res., 32, 2022
|
|
4OCZ
 
 | | Crystal structure of human soluble epoxide hydrolase complexed with 1-(1-isobutyrylpiperidin-4-yl)-3-(4-(trifluoromethyl)phenyl)urea | | Descriptor: | 1-[1-(2-methylpropanoyl)piperidin-4-yl]-3-[4-(trifluoromethyl)phenyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Lee, K.S.S, Liu, J, Wagner, K.M, Pakhomova, S, Dong, H, Morriseau, C, Fu, S.H, Yang, J, Wang, P, Ulu, A, Mate, C, Nguyen, L, Wullf, H, Eldin, M.L, Mara, A.A, Newcomer, M.E, Zeldin, D.C, Hammock, B.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Optimized inhibitors of soluble epoxide hydrolase improve in vitro target residence time and in vivo efficacy.
J.Med.Chem., 57, 2014
|
|
7KKK
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody Nb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KKL
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody mNb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
8IQY
 
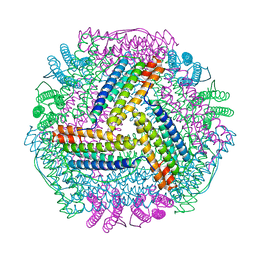 | |
8IQV
 
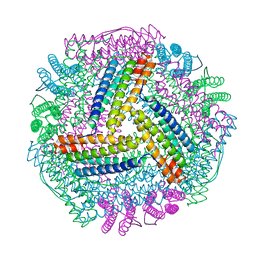 | |
3Q5Y
 
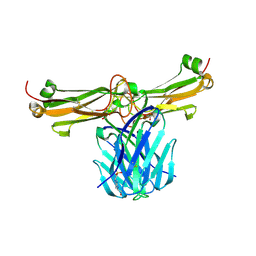 | | V beta/V beta homodimerization-based pre-TCR model suggested by TCR beta crystal structures | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Chen, Q, Zhang, H, Wang, J.-H. | | Deposit date: | 2010-12-30 | | Release date: | 2011-03-09 | | Last modified: | 2014-10-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A conserved hydrophobic patch on Vbeta domains revealed by TCRbeta chain crystal structures: implications for pre-TCR dimerization
Front Immunol, 2, 2011
|
|
7WWP
 
 | | Crystal structure of human Npl4 | | Descriptor: | Nuclear protein localization protein 4 homolog, ZINC ION | | Authors: | Nguyen, T.Q, Le, L.T.M, Kim, D.H, Ko, K.S, Lee, H.T, Nguyen, Y.T.K, Kim, H.S, Han, B.W, Kang, W, Yang, J.K. | | Deposit date: | 2022-02-14 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis for the interaction between human Npl4 and Npl4-binding motif of human Ufd1.
Structure, 30, 2022
|
|
