1MQA
 
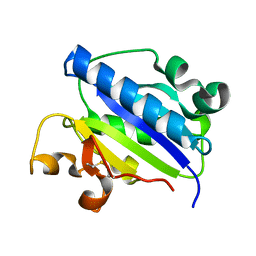 | | Crystal structure of high affinity alphaL I domain in the absence of ligand or metal | | 分子名称: | Integrin alpha-L | | 著者 | Shimaoka, T, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, Zhang, R, Takagi, J, Wang, J.-H, Springer, T.A. | | 登録日 | 2002-09-15 | | 公開日 | 2003-01-14 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
1MQ9
 
 | | Crystal structure of high affinity alphaL I domain with ligand mimetic crystal contact | | 分子名称: | Integrin alpha-L, MANGANESE (II) ION | | 著者 | Shimaoka, M, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, McCormack, A, Zhang, R, Joachimiak, A, Takagi, J, Wang, J.-H, Springer, T.A. | | 登録日 | 2002-09-15 | | 公開日 | 2003-01-14 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
1MJN
 
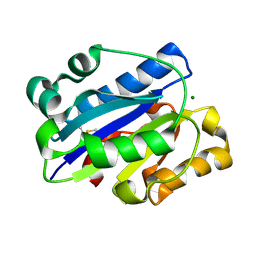 | | Crystal Structure of the intermediate affinity aL I domain mutant | | 分子名称: | Integrin alpha-L, MAGNESIUM ION | | 著者 | Shimaoka, M, Xiao, T, Liu, J.H, Yang, Y.T, Dong, Y.C, Jun, C.D, McCormack, A, Zhang, R.G, Wang, J.H, Springer, T.A. | | 登録日 | 2002-08-28 | | 公開日 | 2003-01-28 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structures of the alphaL I Domain and its Complex with ICAM-1 reveal a Shape-shifting Pathway for Integrin Regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
5VM7
 
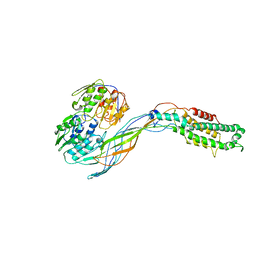 | | Pseudo-atomic model of the MurA-A2 complex | | 分子名称: | Maturation protein A2, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | 著者 | Cui, Z, Zhang, J. | | 登録日 | 2017-04-26 | | 公開日 | 2017-10-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (5.7 Å) | | 主引用文献 | Structures of Q beta virions, virus-like particles, and the Q beta-MurA complex reveal internal coat proteins and the mechanism of host lysis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1VYU
 
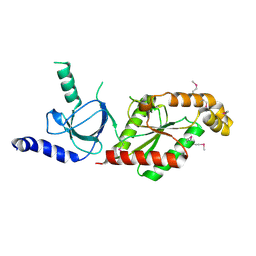 | | Beta3 subunit of Voltage-gated Ca2+-channel | | 分子名称: | CALCIUM CHANNEL BETA-3 SUBUNIT | | 著者 | Chen, Y.-H, Li, M.-H, Zhang, Y, He, L.-L, Yamada, Y, Fitzmaurice, A, Yang, S, Zhang, H, Liang, T, Yang, J. | | 登録日 | 2004-05-07 | | 公開日 | 2004-06-15 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Nature, 429, 2004
|
|
7LTW
 
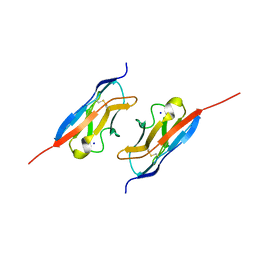 | | Crystal structure of the mouse Kirrel2 D1 homodimer | | 分子名称: | Kin of IRRE-like protein 2, SODIUM ION | | 著者 | Roman, C.A, Pak, J.S, Wang, J, Ozkan, E. | | 登録日 | 2021-02-20 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Molecular and structural basis of olfactory sensory neuron axon coalescence by Kirrel receptors.
Cell Rep, 37, 2021
|
|
1VYV
 
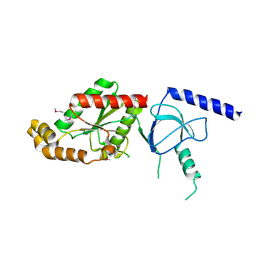 | | beta4 subunit of Ca2+ channel | | 分子名称: | CALCIUM CHANNEL BETA-4SUBUNIT | | 著者 | Chen, Y.-H, Li, M.-H, Zhang, Y, He, L.-L, Yamada, Y, Fitzmaurice, A, Yang, S, Zhang, H, Liang, T, Yang, J. | | 登録日 | 2004-05-07 | | 公開日 | 2004-06-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Nature, 429, 2004
|
|
1MIT
 
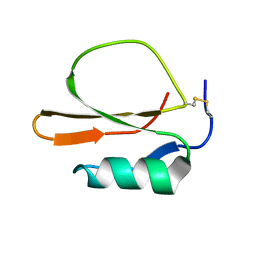 | | RECOMBINANT CUCURBITA MAXIMA TRYPSIN INHIBITOR V (RCMTI-V) (NMR, MINIMIZED AVERAGE STRUCTURE) | | 分子名称: | TRYPSIN INHIBITOR V | | 著者 | Cai, M, Gong, Y, Huang, Y, Liu, J, Prakash, O, Wen, L, Wen, J.J, Huang, J.-K, Krishnamoorthi, R. | | 登録日 | 1995-10-26 | | 公開日 | 1996-04-03 | | 最終更新日 | 2017-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and backbone dynamics of recombinant Cucurbita maxima trypsin inhibitor-V determined by NMR spectroscopy.
Biochemistry, 35, 1996
|
|
5VFK
 
 | |
1VYT
 
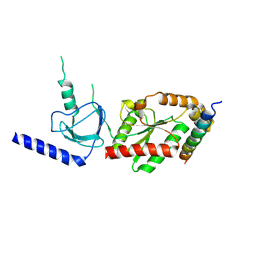 | | beta3 subunit complexed with aid | | 分子名称: | CALCIUM CHANNEL BETA-3 SUBUNIT, VOLTAGE-DEPENDENT L-TYPE CALCIUM CHANNEL ALPHA-1C SUBUNIT | | 著者 | Chen, Y.-H, Li, M.-H, Zhang, Y, He, L.-L, Yamada, Y, Fitzmaurice, A, Yang, S, Zhang, H, Tong, L, Yang, J. | | 登録日 | 2004-05-07 | | 公開日 | 2004-06-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Nature, 429, 2004
|
|
8SSU
 
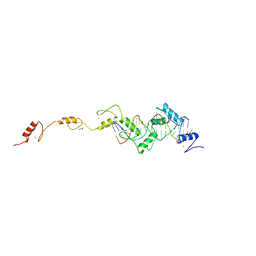 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 19mer DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA (19-MER) Strand I, DNA (19-MER) Strand II, ... | | 著者 | Horton, J.R, Yang, J, Cheng, X. | | 登録日 | 2023-05-08 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
7YK6
 
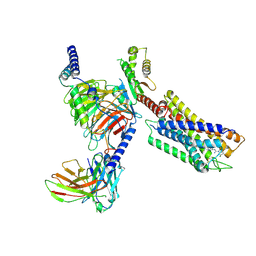 | | Cryo-EM structure of the compound 4-bound human relaxin family peptide receptor 4 (RXFP4)-Gi complex | | 分子名称: | 1-[2-(4-chlorophenyl)ethyl]-3-[(7-ethyl-5-oxidanyl-1H-indol-3-yl)methylideneamino]guanidine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Chen, Y, Zhou, Q.T, Wang, J, Xu, Y.W, Wang, Y, Yan, J.H, Wang, Y.B, Zhu, Q, Zhao, F.H, Li, C.H, Chen, C.W, Cai, X.Q, Bathgate, R.A.D, Shen, C, Liu, H, Xu, H.E, Yang, D.H, Wang, M.W. | | 登録日 | 2022-07-21 | | 公開日 | 2023-03-01 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | Ligand recognition mechanism of the human relaxin family peptide receptor 4 (RXFP4).
Nat Commun, 14, 2023
|
|
7YJ4
 
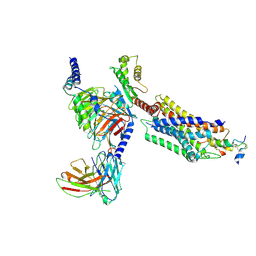 | | Cryo-EM structure of the INSL5-bound human relaxin family peptidereceptor 4 (RXFP4)-Gi complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-2, ... | | 著者 | Chen, Y, Zhou, Q.T, Wang, J, Xu, Y.W, Wang, Y, Yan, J.H, Wang, Y.B, Zhu, Q, Zhao, F.H, Li, C.H, Chen, C.W, Cai, X.Q, Bathgate, R.A.D, Shen, C, Liu, H, Xu, H.E, Yang, D.H, Wang, M.W. | | 登録日 | 2022-07-19 | | 公開日 | 2023-03-01 | | 実験手法 | ELECTRON MICROSCOPY (3.19 Å) | | 主引用文献 | Ligand recognition mechanism of the human relaxin family peptide receptor 4 (RXFP4).
Nat Commun, 14, 2023
|
|
7YK7
 
 | | Cryo-EM structure of the DC591053-bound human relaxin family peptide receptor 4 (RXFP4)-Gi complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-2, ... | | 著者 | Chen, Y, Zhou, Q.T, Wang, J, Xu, Y.W, Wang, Y, Yan, J.H, Wang, Y.B, Zhu, Q, Zhao, F.H, Li, C.H, Chen, C.W, Cai, X.Q, Bathgate, R.A.D, Shen, C, Xu, H.E, Yang, D.H, Liu, H, Wang, M.W. | | 登録日 | 2022-07-21 | | 公開日 | 2023-03-01 | | 実験手法 | ELECTRON MICROSCOPY (2.75 Å) | | 主引用文献 | Ligand recognition mechanism of the human relaxin family peptide receptor 4 (RXFP4).
Nat Commun, 14, 2023
|
|
5KZN
 
 | | Metabotropic Glutamate Receptor | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Metabotropic glutamate receptor 2 | | 著者 | Chappell, M.D, Li, R, Smith, S.C, Dressman, B.A, Tromiczak, E.G, Tripp, A.E, Blanco, M.-J, Vetman, T, Quimby, S.J, Matt, J, Britton, T, Fivush, A.M, Schkeryantz, J.M, Mayhugh, D, Erickson, J.A, Bures, M, Jaramillo, C, Carpintero, M, de Diego, J.E, Barberis, M, Garcia-Cerrada, S, Soriano, J.F, Antonysamy, S, Atwell, S, MacEwan, I, Condon, B, Bradley, C, Wang, J, Zhang, A, Conners, K, Groshong, C, Wasserman, S.R, Koss, J.W, Witkin, J.M, Li, X, Overshiner, C, Wafford, K.A, Seidel, W, Wang, X.-S, Heinz, B.A, Swanson, S, Catlow, J, Bedwell, D, Monn, J.A, Mitch, C.H, Ornstein, P. | | 登録日 | 2016-07-25 | | 公開日 | 2016-12-28 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Discovery of (1S,2R,3S,4S,5R,6R)-2-Amino-3-[(3,4-difluorophenyl)sulfanylmethyl]-4-hydroxy-bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid Hydrochloride (LY3020371HCl): A Potent, Metabotropic Glutamate 2/3 Receptor Antagonist with Antidepressant-Like Activity.
J. Med. Chem., 59, 2016
|
|
6JUI
 
 | | The atypical Myb-like protein Cdc5 contains two distinct nucleic acid-binding surfaces | | 分子名称: | Pre-mRNA-splicing factor CEF1 | | 著者 | Wang, C, Li, G, Li, M, Yang, J, Liu, J. | | 登録日 | 2019-04-14 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.402 Å) | | 主引用文献 | Two distinct nucleic acid binding surfaces of Cdc5 regulate development.
Biochem.J., 476, 2019
|
|
6KHX
 
 | | Crystal structure of Prx from Akkermansia muciniphila | | 分子名称: | CALCIUM ION, Peroxiredoxin | | 著者 | Li, M, Wang, J, Xu, W, Wang, Y, Zhang, M, Wang, M. | | 登録日 | 2019-07-16 | | 公開日 | 2020-02-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Crystal structure of Akkermansia muciniphila peroxiredoxin reveals a novel regulatory mechanism of typical 2-Cys Prxs by a distinct loop.
Febs Lett., 594, 2020
|
|
7LW2
 
 | |
6LAA
 
 | | Crystal structure of full-length CYP116B46 from Tepidiphilus thermophilus | | 分子名称: | 1,2-ETHANEDIOL, CARBONATE ION, Cytochrome P450, ... | | 著者 | Zhang, L.L, Ko, T.P, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | 登録日 | 2019-11-12 | | 公開日 | 2020-06-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Structural insight into the electron transfer pathway of a self-sufficient P450 monooxygenase.
Nat Commun, 11, 2020
|
|
7Y78
 
 | | Crystal structure of Cry78Aa | | 分子名称: | 1,2-ETHANEDIOL, AMMONIUM ION, Toxin | | 著者 | Cao, B.B, Nie, Y.F, Wang, N.C, Guan, Z.Y, Zhang, D.L, Zhang, J. | | 登録日 | 2022-06-21 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The crystal structure of Cry78Aa from Bacillus thuringiensis provides insights into its insecticidal activity.
Commun Biol, 5, 2022
|
|
7XU2
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU0
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-211 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XTZ
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-1 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU1
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-122 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU4
 
 | | Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
