2MUW
 
 | | NOE-based model of the influenza A virus N31S mutant (19-49) bound to drug 11 | | 分子名称: | (3s,5s,7s)-N-[(5-bromothiophen-2-yl)methyl]tricyclo[3.3.1.1~3,7~]decan-1-aminium, Matrix protein 2 | | 著者 | Wu, Y, Wang, J, DeGrado, W. | | 登録日 | 2014-09-18 | | 公開日 | 2014-12-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Flipping in the Pore: Discovery of Dual Inhibitors That Bind in Different Orientations to the Wild-Type versus the Amantadine-Resistant S31N Mutant of the Influenza A Virus M2 Proton Channel.
J.Am.Chem.Soc., 136, 2014
|
|
6F9J
 
 | | Crystal structure of Barley Beta-Amylase complexed with 4-O-alpha-D-mannopyranosyl-(1-deoxynojirimycin) | | 分子名称: | Beta-amylase, CHLORIDE ION, alpha-D-mannopyranose-(1-4)-1-DEOXYNOJIRIMYCIN | | 著者 | Moncayo, M.A, Rodrigues, L.L, Stevenson, C.E.M, Ruzanski, C, Rejzek, M, Lawson, D.M, Angulo, J, Field, R.A. | | 登録日 | 2017-12-14 | | 公開日 | 2019-01-30 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Synthesis, biological and structural analysis of prospective glycosyl-iminosugar prodrugs: impact on germination
To be published
|
|
2Y7B
 
 | | Crystal structure of the PH domain of human Actin-binding protein anillin ANLN | | 分子名称: | 1,2-ETHANEDIOL, ACTIN-BINDING PROTEIN ANILLIN | | 著者 | Vollmar, M, Wang, J, Krojer, T, Elkins, J, Filippakopoulos, P, Ugochukwu, E, Cocking, R, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S. | | 登録日 | 2011-01-31 | | 公開日 | 2011-03-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of the Ph Domain of Human Actin-Binding Protein Anillin Anln
To be Published
|
|
2L7B
 
 | | NMR Structure of full length apoE3 | | 分子名称: | Apolipoprotein E | | 著者 | Chen, J, Wang, J. | | 登録日 | 2010-12-07 | | 公開日 | 2011-08-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4FHK
 
 | | Crystal Structure of PI3K-gamma in Complex with Imidazopyridazine 19e | | 分子名称: | 3-[2-methyl-6-(pyrazin-2-ylamino)pyrimidin-4-yl]-N-(1H-pyrazol-3-yl)imidazo[1,2-b]pyridazin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | 著者 | Shaffer, P.L, Tang, J, Yakowec, P. | | 登録日 | 2012-06-06 | | 公開日 | 2013-04-10 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Discovery and optimization of potent and selective imidazopyridine and imidazopyridazine mTOR inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2LEO
 
 | |
5E8E
 
 | | Crystal structure of thrombin bound to an exosite 1-specific IgA Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | 著者 | Baglin, T.P, Langdown, J, Frasson, R, Huntington, J.A. | | 登録日 | 2015-10-14 | | 公開日 | 2015-10-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery and characterization of an antibody directed against exosite I of thrombin.
J.Thromb.Haemost., 14, 2016
|
|
5EA7
 
 | | Crystal Structure of Inhibitor BMS-433771 in Complex with Prefusion RSV F Glycoprotein | | 分子名称: | 1-cyclopropyl-3-[[1-(4-oxidanylbutyl)benzimidazol-2-yl]methyl]imidazo[4,5-c]pyridin-2-one, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Fusion glycoprotein F0, ... | | 著者 | Battles, M.B, McLellan, J.S, Arnoult, E, Roymans, D, Langedijk, J.P. | | 登録日 | 2015-10-15 | | 公開日 | 2015-12-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.851 Å) | | 主引用文献 | Molecular mechanism of respiratory syncytial virus fusion inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
5V9P
 
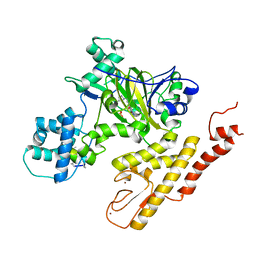 | | Crystal structure of pyrrolidine amide inhibitor [(3S)-3-(4-bromo-1H-pyrazol-1-yl)pyrrolidin-1-yl][3-(propan-2-yl)-1H-pyrazol-5-yl]methanone (compound 35) in complex with KDM5A | | 分子名称: | Lysine-specific demethylase 5A, NICKEL (II) ION, SULFATE ION, ... | | 著者 | Kiefer, J.R, Liang, J, Vinogradova, M. | | 登録日 | 2017-03-23 | | 公開日 | 2017-05-10 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | From a novel HTS hit to potent, selective, and orally bioavailable KDM5 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
4CBT
 
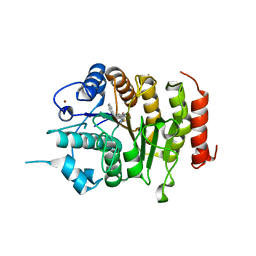 | | Design, synthesis, and biological evaluation of potent and selective Class IIa HDAC inhibitors as a potential therapy for Huntington's disease | | 分子名称: | (1R,2R,3R)-2-[4-(5-fluoranylpyrimidin-2-yl)phenyl]-N-oxidanyl-3-phenyl-cyclopropane-1-carboxamide, HISTONE DEACETYLASE 4, ZINC ION | | 著者 | Burli, R.W, Luckhurst, C.A, Aziz, O, Matthews, K.L, Yates, D, Lyons, K.A, Beconi, M, McAllister, G, Breccia, P, Stott, A.J, Penrose, S.D, Wall, M, Lamers, M, Leonard, P, Mueller, I, Richardson, C.M, Jarvis, R, Stones, L, Hughes, S, Wishart, G, Haughan, A.F, O'Connell, C, Mead, T, McNeil, H, Vann, J, Mangette, J, Maillard, M, Beaumont, V, Munoz-Sanjuan, I, Dominguez, C. | | 登録日 | 2013-10-16 | | 公開日 | 2013-12-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.03 Å) | | 主引用文献 | Design, synthesis, and biological evaluation of potent and selective class IIa histone deacetylase (HDAC) inhibitors as a potential therapy for Huntington's disease.
J. Med. Chem., 56, 2013
|
|
4MPE
 
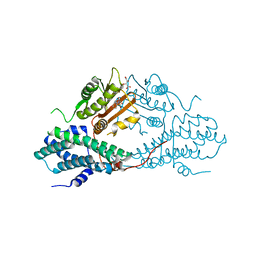 | | Crystal structure of pyruvate dehydrogenase kinase isoform 2 in complex with inhibitor PS8 | | 分子名称: | 4-[(5-hydroxy-1,3-dihydro-2H-isoindol-2-yl)sulfonyl]benzene-1,3-diol, L(+)-TARTARIC ACID, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 2, ... | | 著者 | Gui, W.J, Tso, S.C, Chuang, J.L, Wu, C.Y, Qi, X, Tambar, U.K, Wynn, R.M, Chuang, D.T. | | 登録日 | 2013-09-12 | | 公開日 | 2014-01-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.953 Å) | | 主引用文献 | Structure-guided Development of Specific Pyruvate Dehydrogenase Kinase Inhibitors Targeting the ATP-binding Pocket.
J.Biol.Chem., 289, 2014
|
|
7ER3
 
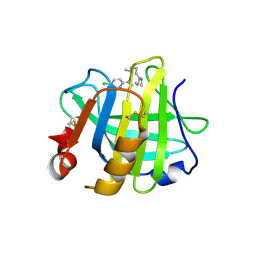 | | Crystal structure of beta-lactoglobulin complexed with chloroquine | | 分子名称: | (4S)-N~4~-(7-chloroquinolin-4-yl)-N~1~,N~1~-diethylpentane-1,4-diamine, Major allergen beta-lactoglobulin | | 著者 | Yao, Q, Ma, J, Xing, Y, Zang, J. | | 登録日 | 2021-05-05 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.598 Å) | | 主引用文献 | Binding of Chloroquine to Whey Protein Relieves Its Cytotoxicity while Enhancing Its Uptake by Cells.
J.Agric.Food Chem., 69, 2021
|
|
5C27
 
 | | Crystal structure of SYK in complex with compound 2 | | 分子名称: | 3-{8-[(3,4-dimethoxyphenyl)amino]imidazo[1,2-a]pyrazin-6-yl}-N-[4-(methylcarbamoyl)phenyl]benzamide, GLU-VAL-TYR-GLU-SER, GLYCEROL, ... | | 著者 | Han, S, Chang, J. | | 登録日 | 2015-06-15 | | 公開日 | 2015-10-07 | | 最終更新日 | 2016-02-03 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Imidazotriazines: Spleen Tyrosine Kinase (Syk) Inhibitors Identified by Free-Energy Perturbation (FEP).
Chemmedchem, 11, 2016
|
|
2JYJ
 
 | | Re-refining the tetraloop-receptor RNA-RNA complex using NMR-derived restraints and Xplor-nih (2.18) | | 分子名称: | RNA (43-MER) | | 著者 | Zuo, X, Wang, J, Foster, T.R, Schwieters, C.D, Tiede, D.M, Butcher, S.E, Wang, Y. | | 登録日 | 2007-12-13 | | 公開日 | 2008-10-07 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | RNA helical packing in solution: NMR structure of a 30 kDa GAAA tetraloop-receptor complex.
J.Mol.Biol., 351, 2005
|
|
6GGY
 
 | | Paenibacillus sp. YM1 laminaribiose phosphorylase with sulphate bound | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Laminaribiose phosphorylase, ... | | 著者 | Kuhaudomlarp, S, Walpole, S, Stevenson, C.E.M, Nepogodiev, S.A, Lawson, D.M, Angulo, J, Field, R.A. | | 登録日 | 2018-05-04 | | 公開日 | 2018-06-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Unravelling the Specificity of Laminaribiose Phosphorylase from Paenibacillus sp. YM-1 towards Donor Substrates Glucose/Mannose 1-Phosphate by Using X-ray Crystallography and Saturation Transfer Difference NMR Spectroscopy.
Chembiochem, 20, 2019
|
|
7KGB
 
 | |
1DEI
 
 | | DESHEPTAPEPTIDE (B24-B30) INSULIN | | 分子名称: | INSULIN | | 著者 | Bao, S.-J, Chang, W.-R, Wan, Z.-L, Zhang, J.-P, Liang, D.-C. | | 登録日 | 1996-05-15 | | 公開日 | 1997-06-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of desheptapeptide(B24-B30)insulin at 1.6 A resolution: implications for receptor binding.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
6GH3
 
 | | Paenibacillus sp. YM1 laminaribiose phosphorylase with alpha-man-1-phosphate bound | | 分子名称: | 1,2-ETHANEDIOL, 1-O-phosphono-alpha-D-mannopyranose, CHLORIDE ION, ... | | 著者 | Kuhaudomlarp, S, Walpole, S, Stevenson, C.E.M, Nepogodiev, S.A, Lawson, D.M, Angulo, J, Field, R.A. | | 登録日 | 2018-05-04 | | 公開日 | 2018-06-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Unravelling the Specificity of Laminaribiose Phosphorylase from Paenibacillus sp. YM-1 towards Donor Substrates Glucose/Mannose 1-Phosphate by Using X-ray Crystallography and Saturation Transfer Difference NMR Spectroscopy.
Chembiochem, 20, 2019
|
|
3C1P
 
 | | Crystal Structure of an alternating D-Alanyl, L-Homoalanyl PNA | | 分子名称: | Peptide Nucleic Acid DLY-HGL-AGD-LHC-AGD-LHC-CUD-LYS | | 著者 | Cuesta-Seijo, J.A, Sheldrick, G.M, Zhang, J, Diederichsen, U. | | 登録日 | 2008-01-23 | | 公開日 | 2009-01-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Continuous beta-turn fold of an alternating alanyl/homoalanyl peptide nucleic acid.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5DOQ
 
 | | The structure of bd oxidase from Geobacillus thermodenitrificans | | 分子名称: | Bd-type quinol oxidase subunit I, Bd-type quinol oxidase subunit II, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, ... | | 著者 | Safarian, S, Mueller, H, Rajendran, C, Preu, J, Ovchinnikov, S, Kusumoto, T, Hirose, T, Langer, J, Sakamoto, J, Michel, H. | | 登録日 | 2015-09-11 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases.
Science, 352, 2016
|
|
4GO1
 
 | | Crystal Structure of full length transcription repressor LsrR from E. coli. | | 分子名称: | GLYCEROL, Transcriptional regulator LsrR | | 著者 | Wu, M, Tao, Y, Liu, X, Zang, J. | | 登録日 | 2012-08-17 | | 公開日 | 2013-04-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Basis for Phosphorylated Autoinducer-2 Modulation of the Oligomerization State of the Global Transcription Regulator LsrR from Escherichia coli
J.Biol.Chem., 288, 2013
|
|
5TSP
 
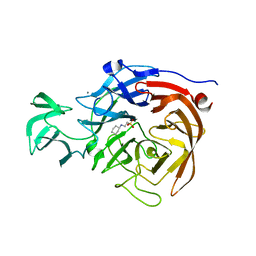 | | Crystal structure of the catalytic domain of Clostridium perfringens neuraminidase (NanI) in complex with a CHES | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, Sialidase | | 著者 | Lee, Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Kang, J.Y, Jin, M.S, Ryu, Y.B, Park, K.H, Eom, S.H. | | 登録日 | 2016-10-31 | | 公開日 | 2017-03-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | Crystal structure of the catalytic domain of Clostridium perfringens neuraminidase in complex with a non-carbohydrate-based inhibitor, 2-(cyclohexylamino)ethanesulfonic acid
Biochem. Biophys. Res. Commun., 486, 2017
|
|
5C26
 
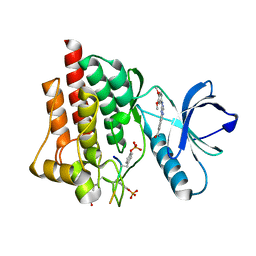 | | Crystal structure of SYK in complex with compound 1 | | 分子名称: | 3-{8-[(3,4-dimethoxyphenyl)amino]imidazo[1,2-a]pyrazin-6-yl}benzamide, GLU-VAL-PTR-GLU-SER-PRO, Tyrosine-protein kinase SYK | | 著者 | Han, S, Chang, J. | | 登録日 | 2015-06-15 | | 公開日 | 2015-10-07 | | 最終更新日 | 2016-02-03 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Imidazotriazines: Spleen Tyrosine Kinase (Syk) Inhibitors Identified by Free-Energy Perturbation (FEP).
Chemmedchem, 11, 2016
|
|
5C6B
 
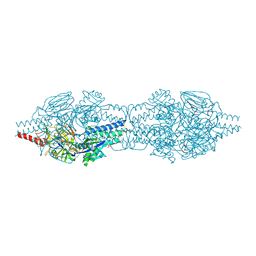 | |
4N19
 
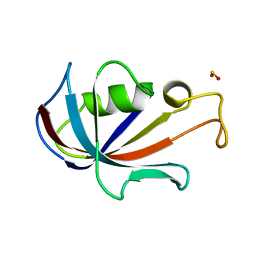 | | Structural basis of conformational transitions in the active site and 80 s loop in the FK506 binding protein FKBP12 | | 分子名称: | Peptidyl-prolyl cis-trans isomerase FKBP1A, SULFATE ION | | 著者 | Mustafi, S.M, Brecher, M.B, Zhang, J, Li, H.M, Lemaster, D.M, Hernandez, G. | | 登録日 | 2013-10-03 | | 公開日 | 2014-02-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structural basis of conformational transitions in the active site and 80's loop in the FK506-binding protein FKBP12.
Biochem.J., 458, 2014
|
|
