5O2S
 
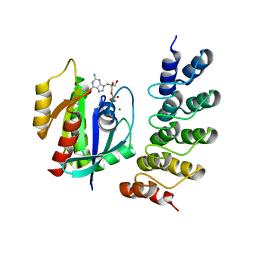 | | Human KRAS in complex with darpin K27 | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Debreczeni, J.E, Guillard, S, Kolasinska-Zwierz, P, Breed, J, Zhang, J, Bery, N, Marwood, R, Tart, J, Stocki, P, Mistry, B, Phillips, C, Rabbitts, T, Jackson, R, Minter, R. | | Deposit date: | 2017-05-22 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural and functional characterization of a DARPin which inhibits Ras nucleotide exchange.
Nat Commun, 8, 2017
|
|
3IZL
 
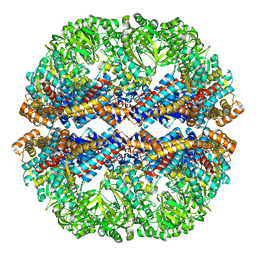 | | Mm-cpn rls deltalid with ATP and AlFx | | Descriptor: | Mm-cpn rls deltalid | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
6AEI
 
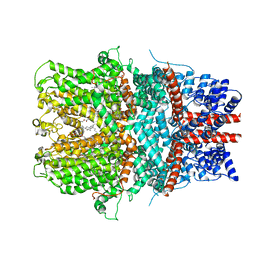 | | Cryo-EM structure of the receptor-activated TRPC5 ion channel | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | Authors: | Duan, J, Li, Z, Li, J, Zhang, J. | | Deposit date: | 2018-08-05 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structure of TRPC5 at 2.8- angstrom resolution reveals unique and conserved structural elements essential for channel function.
Sci Adv, 5, 2019
|
|
3IZN
 
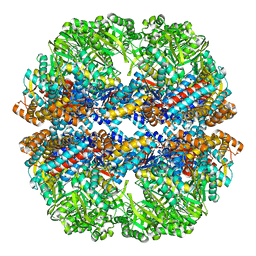 | | Mm-cpn deltalid with ATP | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3IZK
 
 | | Mm-cpn rls deltalid with ATP | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
8XPN
 
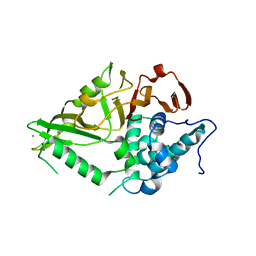 | | The Crystal Structure of USP8 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ubiquitin carboxyl-terminal hydrolase 8, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Wang, J. | | Deposit date: | 2024-01-04 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of USP8 from Biortus.
To Be Published
|
|
6EDJ
 
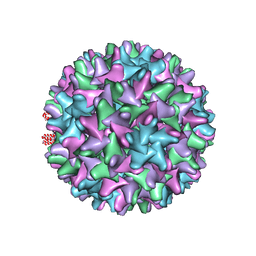 | | Cryo-EM structure of Woodchuck hepatitis virus capsid | | Descriptor: | External core antigen | | Authors: | Zhao, Z, Wang, J.C, Zlotnick, A. | | Deposit date: | 2018-08-09 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.52 Å) | | Cite: | Structural Differences between the Woodchuck Hepatitis Virus Core Protein in the Dimer and Capsid States Are Consistent with Entropic and Conformational Regulation of Assembly.
J.Virol., 93, 2019
|
|
3JTO
 
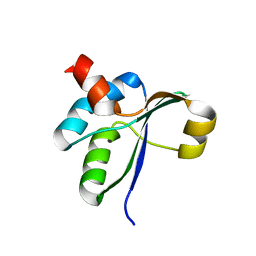 | | Crystal structure of the c-terminal domain of YpbH | | Descriptor: | Adapter protein mecA 2 | | Authors: | Wang, F, Mei, Z, Qi, Y, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the MecA Degradation Tag
To be Published
|
|
8YGZ
 
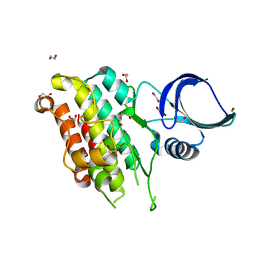 | | The Crystal Structure of TGF beta R2 kinase domain from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, TGF-beta receptor type-2 | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Wang, J. | | Deposit date: | 2024-02-27 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of TGF beta R2 kinase domain from Biortus.
To Be Published
|
|
6LOX
 
 | | Crystal Structure of human glutaminase with macrocyclic inhibitor | | Descriptor: | (E)-15,22-Dioxa-4,11-diaza-5(2,5)-thiadiazola-10(3,6)-pyridazina-1,14(1,3)-dibenzenacyclodocosaphan-18-ene-3,12-dione, Glutaminase kidney isoform, mitochondrial | | Authors: | Bian, J, Li, Z, Xu, X, Wang, J, Li, L. | | Deposit date: | 2020-01-07 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-Enabled Discovery of Novel Macrocyclic Inhibitors Targeting Glutaminase 1 Allosteric Binding Site.
J.Med.Chem., 64, 2021
|
|
8YHP
 
 | | Structure of the PGK1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Phosphoglycerate kinase 1 | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Wang, J. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the PGK1 from Biortus.
To Be Published
|
|
3WDY
 
 | | The complex structure of E113A with cellotetraose | | Descriptor: | Beta-1,3-1,4-glucanase, SULFATE ION, beta-D-glucopyranose, ... | | Authors: | Cheng, Y.S, Huang, C.H, Chen, C.C, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and mutagenetic analyses of a 1,3-1,4-beta-glucanase from Paecilomyces thermophila
Biochim.Biophys.Acta, 1844, 2014
|
|
6ASY
 
 | | BiP-ATP2 | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Liu, Q, Yang, J, Zong, Y, Columbus, L, Zhou, L. | | Deposit date: | 2017-08-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformation transitions of the polypeptide-binding pocket support an active substrate release from Hsp70s.
Nat Commun, 8, 2017
|
|
3J1U
 
 | | Low affinity dynein microtubule binding domain - tubulin complex | | Descriptor: | Cytoplasmic dynein 1 heavy chain 1, seryl t-RNA synthetase chimera, Tubulin alpha-1B chain, ... | | Authors: | Redwine, W.B, Hernandez-Lopez, R, Zou, S, Huang, J, Reck-Peterson, S.L, Leschziner, A.E. | | Deposit date: | 2012-06-25 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Structural basis for microtubule binding and release by dynein.
Science, 337, 2012
|
|
6LXD
 
 | | Pri-miRNA bound DROSHA-DGCR8 complex | | Descriptor: | Microprocessor complex subunit DGCR8, RNA (102-mer), Ribonuclease 3, ... | | Authors: | Jin, W, Wang, J, Liu, C.P, Wang, H.W, Xu, R.M. | | Deposit date: | 2020-02-10 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis for pri-miRNA Recognition by Drosha.
Mol.Cell, 78, 2020
|
|
8XCG
 
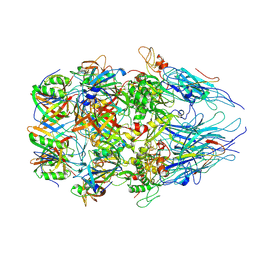 | | Tail tip complex of bacteriophage lambda in the open state | | Descriptor: | IRON/SULFUR CLUSTER, Tail tip assembly protein I, Tail tip protein L, ... | | Authors: | Ge, X.F, Wang, J.W. | | Deposit date: | 2023-12-09 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural mechanism of bacteriophage lambda tail's interaction with the bacterial receptor.
Nat Commun, 15, 2024
|
|
8XCI
 
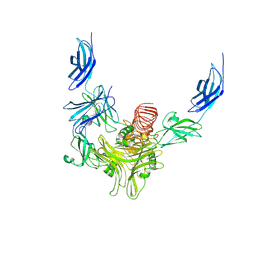 | |
8XCK
 
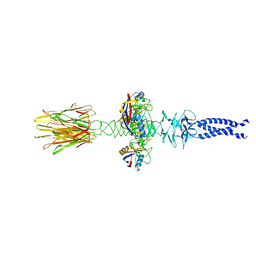 | | Closed state of central tail fiber of bacteriophage lambda | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, Tip attachment protein J | | Authors: | Ge, X.F, Wang, J.W. | | Deposit date: | 2023-12-09 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural mechanism of bacteriophage lambda tail's interaction with the bacterial receptor.
Nat Commun, 15, 2024
|
|
5N8C
 
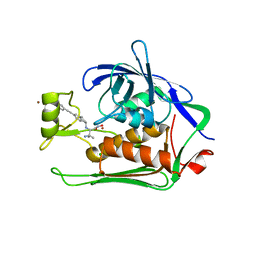 | | Crystal structure of Pseudomonas aeruginosa LpxC complexed with inhibitor | | Descriptor: | (2~{S})-3-azanyl-2-[[(1~{R})-5-[2-[4-[[2-(hydroxymethyl)imidazol-1-yl]methyl]phenyl]ethynyl]-2,3-dihydro-1~{H}-inden-1-yl]amino]-3-methyl-~{N}-oxidanyl-butanamide, CHLORIDE ION, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Cross, J.B, Ryan, M.D, Zhang, J, Cheng, R.K, Wood, M, Andersen, O.A, Brooks, M, Kwong, J, Barker, J. | | Deposit date: | 2017-02-23 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based discovery of LpxC inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
8XCJ
 
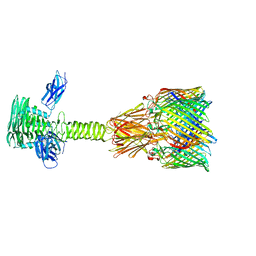 | |
5G2X
 
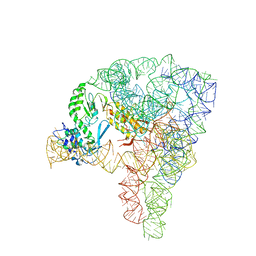 | | Structure a of Group II Intron Complexed with its Reverse Transcriptase | | Descriptor: | 5'-R(*CP*AP*CP*AP*UP*CP*CP*AP*UP*AP*AP*CP)-3', GROUP II INTRON, GROUP II INTRON-ENCODED PROTEIN LTRA | | Authors: | Qu, G, Kaushal, P.S, Wang, J, Shigematsu, H, Piazza, C.L, Agrawal, R.K, Belfort, M, Wang, H.W. | | Deposit date: | 2016-04-16 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of a Group II Intron in Complex with its Reverse Transcriptase.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6AYG
 
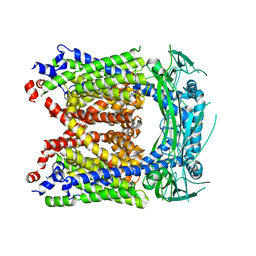 | | Human Apo-TRPML3 channel at pH 4.8 | | Descriptor: | Mucolipin-3 | | Authors: | Zhou, X, Li, M, Su, D, Jia, Q, Li, H, Li, X, Yang, J. | | Deposit date: | 2017-09-08 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.65 Å) | | Cite: | Cryo-EM structures of the human endolysosomal TRPML3 channel in three distinct states.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5ITT
 
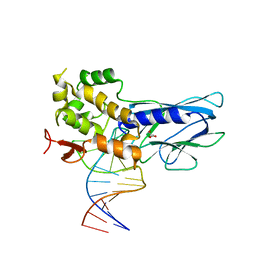 | | Crystal Structure of Human NEIL1 bound to duplex DNA containing THF | | Descriptor: | DNA (26-MER), Endonuclease 8-like 1, GLYCEROL | | Authors: | Zhu, C, Lu, L, Zhang, J, Yue, Z, Song, J, Zong, S, Liu, M, Stovicek, O, Gao, Y, Yi, C. | | Deposit date: | 2016-03-17 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5YKJ
 
 | | Structural basis of the thiol resolving mechanism in yeast mitochondrial 1-Cys peroxiredoxin via glutathione/thioredoxin systems | | Descriptor: | GLYCEROL, Peroxiredoxin PRX1, mitochondrial, ... | | Authors: | Li, C.C, Yang, J, Yang, M.J, Liu, L, Peng, C.T, Li, T, He, L.H, Song, Y.J, Zhu, Y.B, Zhao, N.L, Zhao, C, Bao, R. | | Deposit date: | 2017-10-14 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural basis of the thiol resolving mechanism in yeast mitochondrial 1-Cys peroxiredoxin via glutathione/thioredoxin systems
To be published
|
|
8GHS
 
 | | Empty HBV Cp183 capsid with importin-beta, subparticle reconstruction at 2-fold location | | Descriptor: | Capsid protein | | Authors: | Kim, C, Schlicksup, C.J, Hadden-Perilla, J.A, Wang, J.C.-Y, Zlotnick, A. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the Hepatitis B virus capsid quasi-6-fold with a trapped C-terminal domain reveals capsid movements associated with domain exit.
J.Biol.Chem., 299, 2023
|
|
