4R6I
 
 | | AtxA protein, a virulence regulator from Bacillus anthracis. | | Descriptor: | Anthrax toxin expression trans-acting positive regulator, DODECYL-BETA-D-MALTOSIDE | | Authors: | Osipiuk, J, Horton, L.B, Koehler, T.M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-25 | | Release date: | 2014-10-22 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of Bacillus anthracis virulence regulator AtxA and effects of phosphorylated histidines on multimerization and activity.
Mol.Microbiol., 95, 2015
|
|
4RV8
 
 | | Co-Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Cryptosporidium parvum and the inhibitor p131 | | Descriptor: | 1-(2-{3-[(1E)-N-(2-aminoethoxy)ethanimidoyl]phenyl}propan-2-yl)-3-(4-chloro-3-nitrophenyl)urea, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-11-25 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Structure of Cryptosporidium IMP dehydrogenase bound to an inhibitor with in vivo antiparasitic activity.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
4WJZ
 
 | | Crystal structure of beta-ketoacyl-acyl carrier protein reductase (FabG)(G141A) from Vibrio cholerae | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, PHOSPHATE ION | | Authors: | Hou, J, Zheng, H, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
4WK6
 
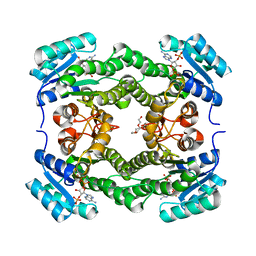 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG) (G141A) from Vibrio cholerae in complex with NADPH | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PENTAETHYLENE GLYCOL | | Authors: | Hou, J, Zheng, H, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
4WH5
 
 | | Crystal structure of lincosamide antibiotic adenylyltransferase LnuA, lincomycin-bound | | Descriptor: | CHLORIDE ION, LINCOMYCIN, Lincosamide resistance protein, ... | | Authors: | Stogios, P.J, Dong, A, Minasov, G, Evdokimova, E, Egorova, O, Kudritska, M, Yim, O, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-20 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | CRYSTAL STRUCTURE OF LINCOSAMIDE ANTIBIOTIC ADENYLYLTRANSFERASE LNUA, LINCOMYCIN BOUND
To Be Published
|
|
4WQK
 
 | | Crystal structure of aminoglycoside nucleotidylyltransferase ANT(2")-Ia, apo form | | Descriptor: | 2''-aminoglycoside nucleotidyltransferase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Cox, G, Stogios, P.J, Savchenko, A, Wright, G.D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Structural and Molecular Basis for Resistance to Aminoglycoside Antibiotics by the Adenylyltransferase ANT(2)-Ia.
Mbio, 6, 2015
|
|
2LOJ
 
 | | Solution NMR structure of TSTM1273 from Salmonella typhimurium LT2, NESG target STT322, CSGID target IDP01027 and OCSP target TSTM1273 | | Descriptor: | Putative cytoplasmic protein | | Authors: | Wu, B, Yee, A, Houliston, S, Garcia, M, Savchenko, A, Arrowsmith, C.H, Anderson, W.F, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of TSTM1273 from Salmonella typhimurium LT2, NESG target STT322, CSGID target IDP01027 and OCSP target TSTM1273
To be Published
|
|
4WQL
 
 | | Crystal structure of aminoglycoside nucleotidylyltransferase ANT(2")-Ia, kanamycin-bound | | Descriptor: | 2''-aminoglycoside nucleotidyltransferase, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Cox, G, Stogios, P.J, Savchenko, A, Wright, G.D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural and Molecular Basis for Resistance to Aminoglycoside Antibiotics by the Adenylyltransferase ANT(2)-Ia.
Mbio, 6, 2015
|
|
4WRR
 
 | | A putative diacylglycerol kinase from Bacillus anthracis str. Sterne | | Descriptor: | MAGNESIUM ION, putative diacylglycerol kinase | | Authors: | Hou, J, Zheng, H, Cooper, D.R, Zimmerman, M.D, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of a putative diacylglycerol kinase from Bacillus anthracis str. Sterne
to be published
|
|
2M46
 
 | | Solution NMR structure of SACOL0876 from Staphylococcus aureus COL, NESG target ZR353 and CSGID target IDP00841 | | Descriptor: | Arsenate reductase, putative | | Authors: | Wu, B, Yee, A, Houliston, S, Garcia, M, Savchenko, A, Arrowsmith, C.H, Anderson, W.F, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of SACOL0876 from Staphylococcus aureus COL, NESG target ZR353 and CSGID target IDP00841
To be Published
|
|
4X9O
 
 | | Beta-ketoacyl-ACP synthase III -2 (FabH2) (C113A) from Vibrio Cholerae soaked with octanoyl-CoA: conformational changes without clearly bound substrate | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 2 | | Authors: | Hou, J, Cooper, D.R, Shabalin, I.G, Grabowski, M, Shumilin, I, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and enzymatic studies of beta-ketoacyl-(acyl carrier protein) synthase III (FabH) from Vibrio cholerae
to be published
|
|
2M4E
 
 | | Solution NMR structure of VV2_0175 from Vibrio vulnificus, NESG target VnR1 and CSGID target IDP91333 | | Descriptor: | Putative uncharacterized protein | | Authors: | Wu, B, Yee, A, Houliston, S, Lemak, A, Garcia, M, Savchenko, A, Arrowsmith, C.H, Anderson, W.F, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-04 | | Release date: | 2013-03-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of VV2_0175 from Vibrio vulnificus, NESG target VnR1 and CSGID target IDP91333
To be Published
|
|
5T07
 
 | | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with Decanoyl-CoA | | Descriptor: | Acyl-CoA thioester hydrolase YbgC, decanoyl-CoA | | Authors: | Watanabe, N, Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-15 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with Decanoyl-CoA
To be published
|
|
5T06
 
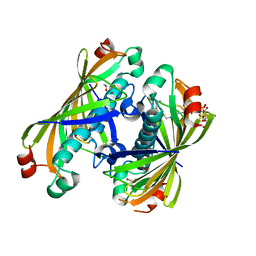 | | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with Hexanoyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, Acyl-CoA thioester hydrolase YbgC, HEXANOYL-COENZYME A | | Authors: | Watanabe, N, Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-15 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with Hexanoyl-CoA
To be published
|
|
5UCC
 
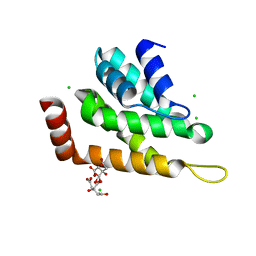 | | Crystal structure of the ENTH domain of ENT2 from Candida albicans | | Descriptor: | CHLORIDE ION, CITRIC ACID, Potential epsin-like clathrin-binding protein | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-22 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the ENTH domain of ENT2 from Candida albicans
To Be Published
|
|
5UC7
 
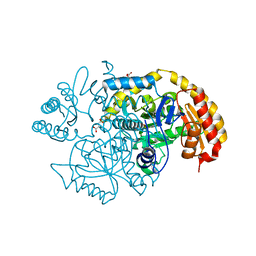 | | Crystal structure of BioA / 7,8-diaminopelargonic acid aminotransferase / DAPA synthase from Citrobacter rodentium, PLP complex | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, D(-)-TARTARIC ACID | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.835 Å) | | Cite: | Crystal structure of BioA / 7,8-diaminopelargonic acid aminotransferase / DAPA synthase from Citrobacter rodentium, PLP complex
To Be Published
|
|
4YFJ
 
 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-Ib | | Descriptor: | Aminoglycoside 3'-N-acetyltransferase, SULFATE ION | | Authors: | Stogios, P.J, Xu, Z, Evdokimova, E, Yim, V, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of aminoglycoside acetyltransferase AAC(3)-Ib
To Be Published
|
|
5U8J
 
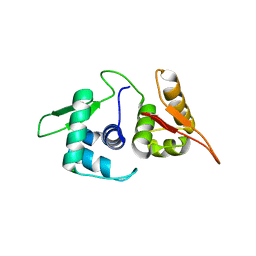 | | Crystal structure of a protein of unknown function ECL_02571 involved in membrane biogenesis from Enterobacter cloacae | | Descriptor: | CHLORIDE ION, UPF0502 protein BFJ73_07745 | | Authors: | Stogios, P.J, Skarina, T, Sandoval, J, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of a protein of unknown function ECL_02571 involved in membrane biogenesis from Enterobacter cloacae
To Be Published
|
|
5UB8
 
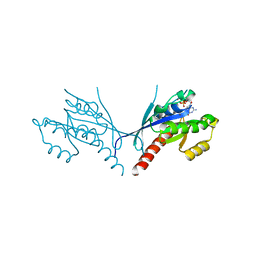 | | Crystal structure of YPT31, a Rab family GTPase from Candida albicans, in complex with GDP and Zn(II) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Likely rab family GTP-binding protein, ZINC ION | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-20 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of YPT31, a Rab family GTPase from Candida albicans, in complex with GDP and Zn(II)
To Be Published
|
|
5UE7
 
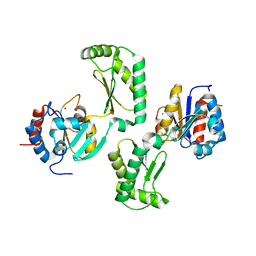 | | Crystal structure of the phosphomannomutase PMM1 from Candida albicans, apoenzyme state | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Phosphomannomutase | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the phosphomannomutase PMM1 from Candida albicans, apoenzyme state
To Be Published
|
|
5U08
 
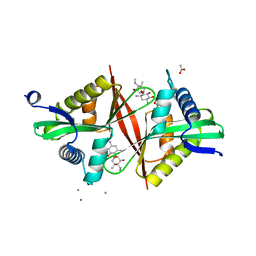 | | Crystal structure of an aminoglycoside acetyltransferase meta-AAC0020 from an uncultured soil metagenomic sample in complex with sisomicin | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, ACETATE ION, CALCIUM ION, ... | | Authors: | Xu, Z, Skarina, T, Wawrzak, Z, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-23 | | Release date: | 2017-02-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes.
ACS Infect Dis, 3, 2017
|
|
5U1H
 
 | | Crystal structure of the C-terminal peptidoglycan binding domain of OprF (PA1777) from Pseudomonas aeruginosa | | Descriptor: | (2R,6S)-2-amino-6-(carboxyamino)-7-{[(1R)-1-carboxyethyl]amino}-7-oxoheptanoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Watanabe, N, Stogios, P.J, Skarina, T, Wawrzak, Z, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-28 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the C-terminal peptidoglycan binding domain of OprF (PA1777) from Pseudomonas aeruginosa
To be published
|
|
5UF8
 
 | | Crystal structure of the ARF family small GTPase ARF2 from Candida albicans in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Potential ADP-ribosylation factor | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-03 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.872 Å) | | Cite: | Crystal structure of the ARF family small GTPase ARF2 from Candida albicans in complex with GDP
To Be Published
|
|
5UXD
 
 | | Crystal structure of macrolide 2'-phosphotransferase MphH from Brachybacterium faecium in complex with azithromycin | | Descriptor: | AZITHROMYCIN, CHLORIDE ION, Macrolide 2'-phosphotransferase MphH, ... | | Authors: | Stogios, P.J, Skarina, T, Wawrzak, Z, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
5UXA
 
 | | Crystal structure of macrolide 2'-phosphotransferase MphB from Escherichia coli | | Descriptor: | CALCIUM ION, Macrolide 2'-phosphotransferase II | | Authors: | Stogios, P.J, Evdokimova, E, Egorova, O, Di Leo, R, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
