8PMD
 
 | | Nucleotide-bound BSEP in nanodiscs | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bile salt export pump, MAGNESIUM ION | | Authors: | Liu, H, Irobalieva, R.N, Kowal, J, Ni, D, Nosol, K, Bang-Sorensen, R, Lancien, L, Stahlberg, H, Stieger, B, Locher, K.P. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of bile salt extrusion and small-molecule inhibition in human BSEP.
Nat Commun, 14, 2023
|
|
8PMJ
 
 | | Vanadate-trapped BSEP in nanodiscs | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Bile salt export pump, ... | | Authors: | Liu, H, Irobalieva, R.N, Kowal, J, Ni, D, Nosol, K, Bang-Sorensen, R, Lancien, L, Stahlberg, H, Stieger, B, Locher, K.P. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structural basis of bile salt extrusion and small-molecule inhibition in human BSEP.
Nat Commun, 14, 2023
|
|
8RQF
 
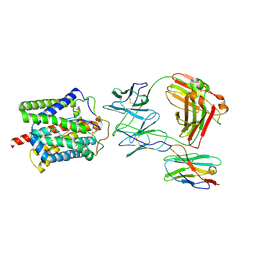 | | Cryo-EM structure of human NTCP-Bulevirtide complex | | Descriptor: | Fab-specific nanobody, Heavy chain of Fab3, Light chain of Fab3, ... | | Authors: | Liu, H, Zakrzewicz, D, Nosol, K, Irobalieva, R.N, Mukherjee, S, Bang-Soerensen, R, Goldmann, N, Kunz, S, Rossi, L, Kossiakoff, A.A, Urban, S, Glebe, D, Geyer, J, Locher, K.P. | | Deposit date: | 2024-01-18 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structure of antiviral drug bulevirtide bound to hepatitis B and D virus receptor protein NTCP.
Nat Commun, 15, 2024
|
|
5L2X
 
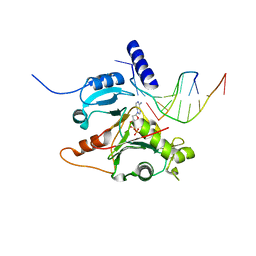 | | Crystal structure of human PrimPol ternary complex | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(P*GP*GP*TP*AP*GP*CP*(DDG))-3'), ... | | Authors: | Rechkoblit, O, Gupta, Y.K, Malik, R, Rajashankar, K.R, Johnson, R.E, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2016-08-02 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and mechanism of human PrimPol, a DNA polymerase with primase activity.
Sci Adv, 2, 2016
|
|
6QUK
 
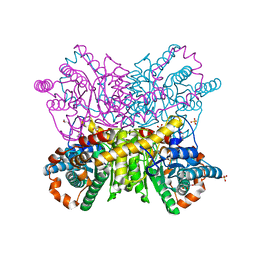 | | Protein crystallization by ionic liquid hydrogel support: glucose isomerase grown by using ionic liquid hydrogel | | Descriptor: | GLYCEROL, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Belviso, B.D, Caliandro, R, Caliandro, R. | | Deposit date: | 2019-02-27 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Protein Crystallization in Ionic-Liquid Hydrogel Composite Membranes
Crystals, 9, 2019
|
|
6QUF
 
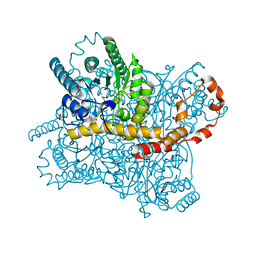 | | Protein crystallization by ionic liquid hydrogel support: reference crystal of glucose isomerase grown on standard silanized glass | | Descriptor: | GLYCEROL, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Belviso, B.D, Caliandro, R, Caliandro, R. | | Deposit date: | 2019-02-27 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Protein Crystallization in Ionic-Liquid Hydrogel Composite Membranes
Crystals, 9, 2019
|
|
3N30
 
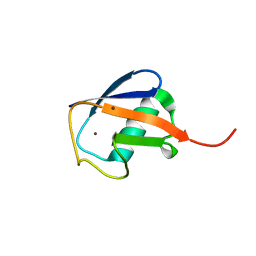 | | Crystal Structure of cubic Zn3-hUb (human ubiquitin) adduct | | Descriptor: | Ubiquitin, ZINC ION | | Authors: | Siliqi, D, Caliandro, R, Arnesano, F, Natile, G, Falini, G, Fermani, S, Belviso, B.D. | | Deposit date: | 2010-05-19 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic analysis of metal-ion binding to human ubiquitin.
Chemistry, 17, 2011
|
|
3N32
 
 | | The crystal structure of human Ubiquitin adduct with Zeise's salt | | Descriptor: | PLATINUM (II) ION, Ubiquitin | | Authors: | Siliqi, D, Caliandro, R, Falini, G, Fermani, S, Natile, G, Arnesano, F, Belviso, B.D. | | Deposit date: | 2010-05-19 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Crystallographic analysis of metal-ion binding to human ubiquitin.
Chemistry, 17, 2011
|
|
7ZLJ
 
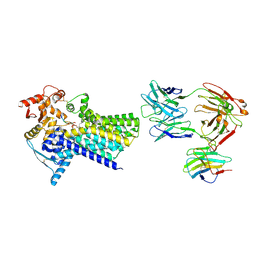 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in ternary complex with Dol25-P-C-Man and acceptor peptide, bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mao, R, Mukherjee, S, Boilevin, J, Irobalieva, R, Darbre, T, Reymond, J.L, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
8AGE
 
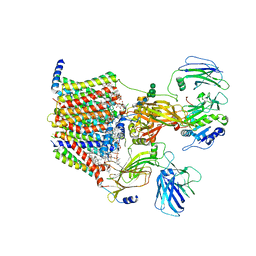 | | Structure of yeast oligosaccharylransferase complex with acceptor peptide bound | | Descriptor: | (2~{S},3~{R},4~{R},5~{S},6~{S})-2-(hydroxymethyl)-6-[(1~{S},2~{R},3~{R},4~{R},5'~{S},6~{S},7~{R},8~{S},9~{R},12~{R},13~{R},15~{S},16~{S},18~{R})-5',7,9,13-tetramethyl-3,15-bis(oxidanyl)spiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icosane-6,2'-oxane]-16-yl]oxy-oxane-3,4,5-triol, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-[3,6-bis(dimethylamino)xanthen-9-yl]-5-methanoyl-benzoate, ... | | Authors: | Ramirez, A.S, de Capitani, M, Pesciullesi, G, Kowal, J, Bloch, J.S, Irobalieva, R.N, Aebi, M, Reymond, J.L, Locher, K.P. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular basis for glycan recognition and reaction priming of eukaryotic oligosaccharyltransferase.
Nat Commun, 13, 2022
|
|
6MRD
 
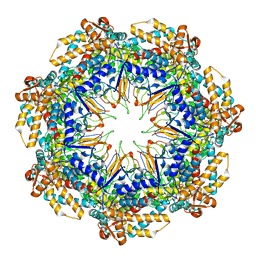 | | ADP-bound human mitochondrial Hsp60-Hsp10 half-football complex | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Gomez-Llorente, Y, Jebara, F, Patra, M, Malik, R, Nissemblat, S, Azem, A, Hirsch, J.A, Ubarretxena-Belandia, I. | | Deposit date: | 2018-10-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis for active single and double ring complexes in human mitochondrial Hsp60-Hsp10 chaperonin.
Nat Commun, 11, 2020
|
|
6MRC
 
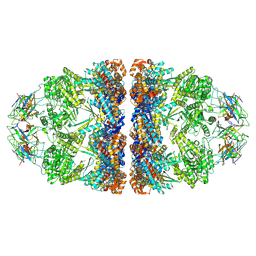 | | ADP-bound human mitochondrial Hsp60-Hsp10 football complex | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Gomez-Llorente, Y, Jebara, F, Patra, M, Malik, R, Nissemblat, S, Azem, A, Hirsch, J.A, Ubarretxena-Belandia, I. | | Deposit date: | 2018-10-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis for active single and double ring complexes in human mitochondrial Hsp60-Hsp10 chaperonin.
Nat Commun, 11, 2020
|
|
7ZYI
 
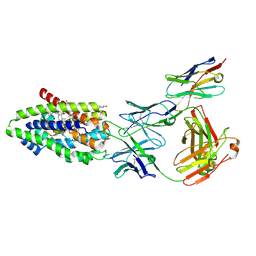 | | Structure of the human sodium/bile acid cotransporter (NTCP) in complex with Fab and nanobody | | Descriptor: | CHOLESTEROL, GLYCOCHENODEOXYCHOLIC ACID, Nanobody, ... | | Authors: | Liu, H, Irobalieva, R.N, Bang-Sorensen, R, Nosol, K, Mukherjee, S, Agrawal, P, Stieger, B, Kossiakoff, A.A, Locher, K.P. | | Deposit date: | 2022-05-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structure of human NTCP reveals the basis of recognition and sodium-driven transport of bile salts into the liver.
Cell Res., 32, 2022
|
|
4P7E
 
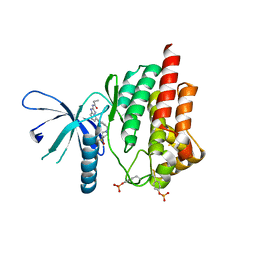 | | Triazolopyridine compounds as selective JAK1 inhibitors: from hit identification to GLPG0634 | | Descriptor: | N-(5-{4-[(1,1-dioxidothiomorpholin-4-yl)methyl]phenyl}[1,2,4]triazolo[1,5-a]pyridin-2-yl)cyclopropanecarboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Menet, C.C.J, Fletcher, S, Van Lommen, G, Geney, R, Blanc, J, Smits, K, Jouannigot, N, van der Aar, E.M, Clement-Lacroix, P, Lepescheux, L, Galien, R, Vayssiere, B, Nelles, L, Christophe, T, Brys, R, Uhring, M, Ciesielski, F, Van Rompaey, L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-11-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Triazolopyridines as Selective JAK1 Inhibitors: From Hit Identification to GLPG0634.
J.Med.Chem., 57, 2014
|
|
7ZLH
 
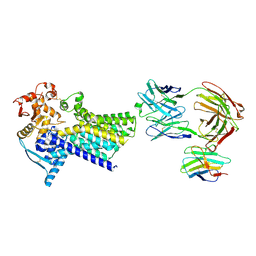 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in apo state, bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mukherjee, S, Irobalieva, R, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
7ZLI
 
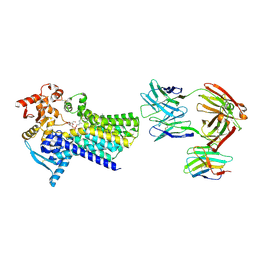 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in complex with Dol25-P-Man and bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mukherjee, S, Boilevin, J, Irobalieva, R, Darbre, T, Reymond, J.L, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
7ZLG
 
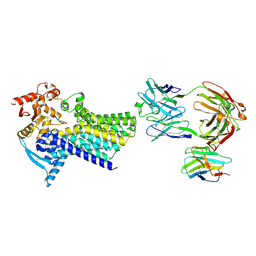 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in complex with acceptor peptide and bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mukherjee, S, Mao, R, Irobalieva, R, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
8AGB
 
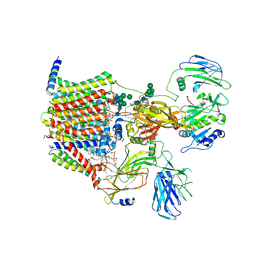 | | Structure of yeast oligosaccharylransferase complex with lipid-linked oligosaccharide bound | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ramirez, A.S, de Capitani, M, Pesciullesi, G, Kowal, J, Bloch, J.S, Irobalieva, R.N, Aebi, M, Reymond, J.L, Locher, K.P. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for glycan recognition and reaction priming of eukaryotic oligosaccharyltransferase.
Nat Commun, 13, 2022
|
|
8AGC
 
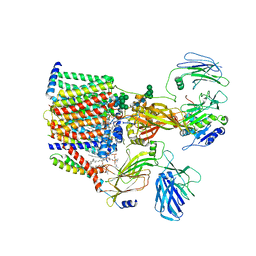 | | Structure of yeast oligosaccharylransferase complex with lipid-linked oligosaccharide and non-acceptor peptide bound | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-[3,6-bis(dimethylamino)xanthen-9-yl]-5-methanoyl-benzoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ramirez, A.S, de Capitani, M, Pesciullesi, G, Kowal, J, Bloch, J.S, Irobalieva, R.N, Aebi, M, Reymond, J.L, Locher, K.P. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for glycan recognition and reaction priming of eukaryotic oligosaccharyltransferase.
Nat Commun, 13, 2022
|
|
4QT5
 
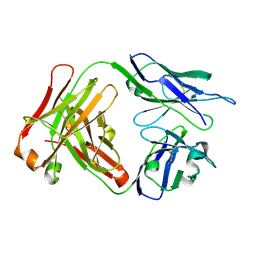 | | Crystal Structure of 3BD10: A Monoclonal Antibody against the TSH Receptor | | Descriptor: | 3BD10 mouse monoclonal antibody, heavy chain, light chain | | Authors: | Hubbard, P.A, Chen, C.R, McLachlan, S.M, Rapoport, B, Murali, R. | | Deposit date: | 2014-07-07 | | Release date: | 2015-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a TSH receptor monoclonal antibody: insight into Graves' disease pathogenesis.
Mol.Endocrinol., 29, 2015
|
|
4QOT
 
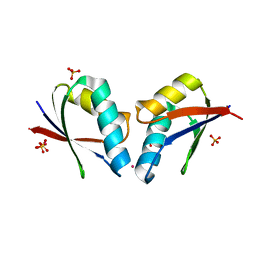 | | Crystal structure of human copper chaperone bound to the platinum ion | | Descriptor: | Copper transport protein ATOX1, PLATINUM (II) ION, SULFATE ION | | Authors: | Belviso, B.D, Galliani, A, Caliandro, R, Arnesano, F, Natile, G. | | Deposit date: | 2014-06-20 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oxaliplatin Binding to Human Copper Chaperone Atox1 and Protein Dimerization
Inorg.Chem., 55, 2016
|
|
6EO8
 
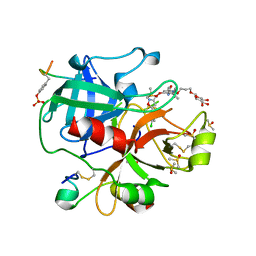 | | Crystal structure of thrombin in complex with a novel glucose-conjugated potent inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, Hirudin variant-2, N-(2-{[5-(5-chlorothiophen-2-yl)-1,2-oxazol-3-yl]methoxy}-6-[3-(beta-D-glucopyranosyloxy)propoxy]phenyl)-1-(propan-2-yl)piperidine-4-carboxamide, ... | | Authors: | Belviso, B.D, Caliandro, R, Aresta, B.M, De Candia, M, Altomare, C.D. | | Deposit date: | 2017-10-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | How a beta-D-glucoside side chain enhances binding affinity to thrombin of inhibitors bearing 2-chlorothiophene as P1 moiety: crystallography, fragment deconstruction study, and evaluation of antithrombotic properties.
J. Med. Chem., 57, 2014
|
|
8FBZ
 
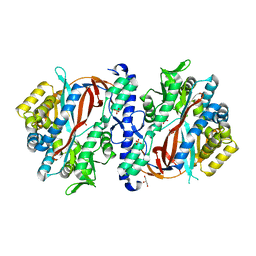 | | Crystal Structure of apo human Glutathione Synthetase Y270E | | Descriptor: | GLYCEROL, Glutathione synthetase, SULFATE ION | | Authors: | Stanford, S.M, Santelli, E, Sankaran, B, Murali, R, Bottini, N. | | Deposit date: | 2022-11-30 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Targeting prostate tumor low-molecular weight tyrosine phosphatase for oxidation-sensitizing therapy.
Sci Adv, 10, 2024
|
|
6EO9
 
 | | Crystal structure of thrombin in complex with a novel glucose-conjugated potent inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, Hirudin variant-2, N-(2-{[5-(5-chlorothiophen-2-yl)-1,2-oxazol-3-yl]methoxy}-6-{3-[(2,3,4,6-tetra-O-acetyl-beta-D-glucopyranosyl)oxy]propoxy}phenyl)-1-(propan-2-yl)piperidine-4-carboxamide, ... | | Authors: | Belviso, B.D, Caliandro, R, Aresta, B.M, De Candia, M, Altomare, C.D. | | Deposit date: | 2017-10-09 | | Release date: | 2017-12-13 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | How a beta-D-glucoside side chain enhances binding affinity to thrombin of inhibitors bearing 2-chlorothiophene as P1 moiety: crystallography, fragment deconstruction study, and evaluation of antithrombotic properties.
J. Med. Chem., 57, 2014
|
|
6RV5
 
 | | X-ray structure of the levansucrase from Erwinia tasmaniensis in complex with levanbiose | | Descriptor: | GLYCEROL, Levansucrase (Beta-D-fructofuranosyl transferase), ZINC ION, ... | | Authors: | Polsinelli, I, Caliandro, R, Demitri, N, Benini, S. | | Deposit date: | 2019-05-31 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The Structure of Sucrose-Soaked Levansucrase Crystals fromErwinia tasmaniensisreveals a Binding Pocket for Levanbiose.
Int J Mol Sci, 21, 2019
|
|
