3P23
 
 | |
2YMJ
 
 | |
2CGE
 
 | |
2CG9
 
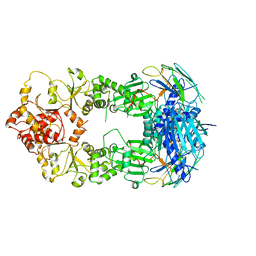 | | Crystal structure of an Hsp90-Sba1 closed chaperone complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, CO-CHAPERONE PROTEIN SBA1 | | Authors: | Ali, M.M.U, Roe, S.M, Prodromou, C, Pearl, L.H. | | Deposit date: | 2006-03-01 | | Release date: | 2006-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of an Hsp90-Nucleotide-P23/Sba1 Closed Chaperone Complex
Nature, 440, 2006
|
|
8T33
 
 | | Crystal structure of K46 acetylated GABARAP in complex with the LIR of TP53INP2/DOR | | Descriptor: | ACETATE ION, Gamma-aminobutyric acid receptor-associated protein, Tumor protein p53-inducible nuclear protein 2, ... | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 20, 2024
|
|
4K89
 
 | | Crystal structure of Pseudomonas aeruginosa strain K solvent tolerant elastase | | Descriptor: | CALCIUM ION, GLYCEROL, Organic solvent tolerant elastase, ... | | Authors: | Ali, M.S.M, Said, Z.S.A.M, Rahman, R.N.Z.R.A, Basri, M, Salleh, A.B. | | Deposit date: | 2013-04-18 | | Release date: | 2014-05-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal structure analysia of solvent tolerant elastase strain K
To be Published
|
|
8T32
 
 | | Crystal structure of K48 acetylated GABARAP in complex with the LIR of TP53INP2/DOR | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein, LIR of DOR, TRIETHYLENE GLYCOL | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 20, 2024
|
|
8T35
 
 | | Crystal structure of K51 acetylated LC3A in complex with the LIR of TP53INP2/DOR | | Descriptor: | 1,2-ETHANEDIOL, Tumor protein p53-inducible nuclear protein 2,Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 20, 2024
|
|
8T36
 
 | | Crystal structure of K49 acetylated LC3A in complex with the LIR of TP53INP2/DOR | | Descriptor: | 1,2-ETHANEDIOL, Tumor protein p53-inducible nuclear protein 2,Microtubule-associated proteins 1A/1B light chain 3A chimera | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 20, 2024
|
|
8T31
 
 | | Crystal structure of GABARAP in complex with the LIR of TP53INP2/DOR | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein, Tumor protein p53-inducible nuclear protein 2 | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 20, 2024
|
|
8T4T
 
 | | Crystal structure of LC3A in complex with the LIR of TP53INP2/DOR | | Descriptor: | Tumor protein p53-inducible nuclear protein 2,Microtubule-associated proteins 1A/1B light chain 3A chimera | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-10 | | Release date: | 2024-05-22 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 20, 2024
|
|
1XOF
 
 | | Heterooligomeric Beta Beta Alpha Miniprotein | | Descriptor: | BBAhetT1 | | Authors: | Ali, M.H, Taylor, C.M, Grigoryan, G, Allen, K.N, Imperiali, B, Keating, A.E. | | Deposit date: | 2004-10-06 | | Release date: | 2005-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design of a Heterospecific, Tetrameric, 21-Residue Miniprotein with Mixed alpha/beta Structure.
Structure, 13, 2005
|
|
8T2N
 
 | | Crystal structure of GABARAP in complex with the LIR of NSs3 | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein, Non-structural protein S | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-06 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | An LIR motif in the Rift Valley fever virus NSs protein is critical for the interaction with LC3 family members and inhibition of autophagy.
Plos Pathog., 20, 2024
|
|
8T2L
 
 | | Crystal structure of LC3A in complex with the LIR of NSs4 | | Descriptor: | Non-structural protein S,Microtubule-associated proteins 1A/1B light chain 3A chimera | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-06 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | An LIR motif in the Rift Valley fever virus NSs protein is critical for the interaction with LC3 family members and inhibition of autophagy.
Plos Pathog., 20, 2024
|
|
8T2M
 
 | | Crystal structure of GABARAP in complex with the LIR of NSs4 | | Descriptor: | Non-structural protein S,Gamma-aminobutyric acid receptor-associated protein chimera | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-06 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | An LIR motif in the Rift Valley fever virus NSs protein is critical for the interaction with LC3 family members and inhibition of autophagy.
Plos Pathog., 20, 2024
|
|
1SN9
 
 | |
1SNA
 
 | | An Oligomeric Domain-Swapped Beta-Beta-Alpha Mini-Protein | | Descriptor: | ISOPROPYL ALCOHOL, tetrameric beta-beta-alpha mini-protein | | Authors: | Ali, M.H, Peisach, E, Allen, K.N, Imperiali, B. | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray structure analysis of a designed oligomeric miniprotein reveals a discrete quaternary architecture.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1SNE
 
 | | An Oligomeric Domain-Swapped Beta-Beta-Alpha Mini-Protein | | Descriptor: | ISOPROPYL ALCOHOL, tetrameric beta-beta-alpha mini-protein | | Authors: | Ali, M.H, Peisach, E, Allen, K.N, Imperiali, B. | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray structure analysis of a designed oligomeric miniprotein reveals a discrete quaternary architecture.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
5ER5
 
 | | Crystal Structure of Calcium-loaded S100B bound to SC1990 | | Descriptor: | CALCIUM ION, ETHIDIUM, Protein S100-B | | Authors: | Cavalier, M.C, Melville, Z.E, Aligholizadeh, E, Fang, L, Alasady, M.J, Pierce, A.D, Wilder, P.T, MacKerell Jr, A.D, Weber, D.J. | | Deposit date: | 2015-11-13 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Novel protein-inhibitor interactions in site 3 of Ca(2+)-bound S100B as discovered by X-ray crystallography.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5ER4
 
 | | Crystal Structure of Calcium-loaded S100B bound to SC0025 | | Descriptor: | 6-methyl-5,6,6~{a},7-tetrahydro-4~{H}-dibenzo[de,g]quinoline-10,11-diol, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Melville, Z.E, Aligholizadeh, E, Fang, L, Alasady, M.J, Pierce, A.D, Wilder, P.T, MacKerell Jr, A.D, Weber, D.J. | | Deposit date: | 2015-11-13 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.813 Å) | | Cite: | Novel protein-inhibitor interactions in site 3 of Ca(2+)-bound S100B as discovered by X-ray crystallography.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5EU7
 
 | | Crystal structure of HIV-1 integrase catalytic core in complex with Fab | | Descriptor: | FAB Heavy Chain, FAB light chain, Integrase | | Authors: | Galilee, M, Griner, S.L, Stroud, R.M, Alian, A. | | Deposit date: | 2015-11-18 | | Release date: | 2016-09-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The Preserved HTH-Docking Cleft of HIV-1 Integrase Is Functionally Critical.
Structure, 24, 2016
|
|
7VLJ
 
 | | Crystal structure of Entamoeba histolytica serine protease inhibitor, Histopin, in the cleaved conformation | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, POTASSIUM ION, ... | | Authors: | Ali, M.F, Devi, S, Gourinath, S. | | Deposit date: | 2021-10-03 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of Entamoeba histolytica serine protease inhibitor, Histopin, in the cleaved conformation
To Be Published
|
|
1KVW
 
 | |
2BPP
 
 | |
6EX9
 
 | |
