9EOQ
 
 | | Cryo-EM Structure of a 1033 Scaffold Base DNA Origami Nanostructure V4 and TBA | | Descriptor: | DNA (42-MER), DNA (5'-D(P*AP*TP*AP*TP*AP*GP*CP*GP*TP*GP*GP*AP*AP*GP*T)-3') | | Authors: | Ali, K, Georg, K, Volodymyr, M, Johanna, G, Maximilian, N.H, Lukas, K, Simone, C, Hendrik, D. | | Deposit date: | 2024-03-15 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Designing Rigid DNA Origami Templates for Molecular Visualization Using Cryo-EM.
Nano Lett., 24, 2024
|
|
6VZS
 
 | | Engineered TTLL6 mutant bound to gamma-elongation analog | | Descriptor: | (2~{S})-2-[[[(3~{R})-3-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VZW
 
 | | TTLL6 bound to the initiation analog | | Descriptor: | (2~{S})-2-[[[(3~{R})-3-acetamido-4-(ethylamino)-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.L, Tanner, T.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VZQ
 
 | | Engineered TTLL6 mutant bound to alpha-elongation analog | | Descriptor: | (2~{S})-2-[[[(1~{R})-1-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, (2~{S})-2-[[[(1~{S})-1-acetamidoethyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, E.K, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VZT
 
 | | TTLL6 bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VZV
 
 | | TTLL6 bound to gamma-elongation analog | | Descriptor: | (2~{S})-2-[[[(3~{R})-3-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, (2~{S})-2-[[[(3~{S})-3-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-oxidanyl-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VZR
 
 | | Engineered TTLL6 bound to the initiation analog | | Descriptor: | (2~{S})-2-[[[(3~{R})-3-acetamido-4-(ethylamino)-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VZU
 
 | | TTLL6 bound to alpha-elongation analog | | Descriptor: | (2~{S})-2-[[[(1~{R})-1-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-oxidanyl-phosphoryl]methyl]pentanedioic acid, (2~{S})-2-[[[(1~{R})-1-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.B, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8T42
 
 | | Model of TTLL6 MTBH1-2 bound to microtubule | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Mahalingan, K.K, Grotjahn, D, Li, Y, Lander, G.C, Zehr, E.A, Roll-Mecak, A. | | Deposit date: | 2023-06-08 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for alpha-tubulin-specific and modification state-dependent glutamylation.
Nat.Chem.Biol., 20, 2024
|
|
8U3Z
 
 | | Model of TTLL6 bound to microtubule from composite map | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Mahalingan, K.K, Grotjahn, D, Li, Y, Lander, G.C, Zehr, E.A, Roll-Mecak, A. | | Deposit date: | 2023-09-08 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for alpha-tubulin-specific and modification state-dependent glutamylation.
Nat.Chem.Biol., 20, 2024
|
|
4IOV
 
 | | The structure of AAVrh32.33, a Novel Gene Delivery Vector | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Capsid protein VP1 | | Authors: | Mikalism, K, Nam, H.-J, Van vilet, K, Vandenberghe, L.H, Mays, L.E, McKenna, R, Wilson, J, Agbandje-McKenna, M. | | Deposit date: | 2013-01-08 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of AAVrh32.33, a novel gene delivery vector.
J.Struct.Biol., 186, 2014
|
|
5VNC
 
 | |
5UW0
 
 | |
5UW1
 
 | |
5UW4
 
 | |
5UX7
 
 | |
5SUL
 
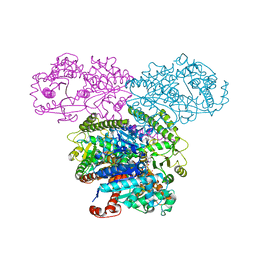 | | Inhibited state structure of yGsy2p | | Descriptor: | Glycogen [starch] synthase isoform 2, URIDINE-5'-DIPHOSPHATE, URIDINE-5'-MONOPHOSPHATE | | Authors: | Mahalingan, K.K, Hurley, T.D. | | Deposit date: | 2016-08-03 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Redox Switch for the Inhibited State of Yeast Glycogen Synthase Mimics Regulation by Phosphorylation.
Biochemistry, 56, 2017
|
|
5MYA
 
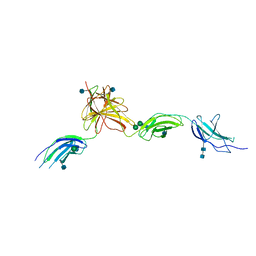 | | Homodimerization of Tie2 Fibronectin-like domains 1-3 in space group C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiopoietin-1 receptor, ... | | Authors: | Leppanen, V.-M, Saharinen, P, Alitalo, K. | | Deposit date: | 2017-01-26 | | Release date: | 2017-04-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of Tie2 activation and Tie2/Tie1 heterodimerization.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MYB
 
 | | Homodimerization of Tie2 Fibronectin-like domains 2 and 3 in space group P21 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiopoietin-1 receptor, ... | | Authors: | Leppanen, V.-M, Saharinen, P, Alitalo, K. | | Deposit date: | 2017-01-26 | | Release date: | 2017-04-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of Tie2 activation and Tie2/Tie1 heterodimerization.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1IZI
 
 | | Inhibitor of HIV protease with unusual binding mode potently inhibiting multi-resistant protease mutants | | Descriptor: | CHLORIDE ION, proteinase, {(1S)-1-BENZYL-4-[3-CARBAMOYL-1-(1-CARBAMOYL-2-PHENYL-ETHYLCARBAMOYL)-(S)-PROPYLCARBAMOYL]-2-OXO-5-PHENYL-PENTYL}-CARBAMIC ACID TERT-BUTYL ESTER | | Authors: | Weber, J, Mesters, J.R, Lepsik, M, Prejdova, J, Svec, M, Sponarova, J, Mlcochova, P, Skalicka, K, Strisovsky, K, Uhlikova, T, Soucek, M, Machala, L, Stankova, M, Vondrasek, J, Klimkait, T, Kraeusslich, H.-G, Hilgenfeld, R, Konvalinka, J. | | Deposit date: | 2002-10-02 | | Release date: | 2002-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Unusual Binding Mode of an HIV-1 Protease Inhibitor Explains its Potency against Multi-drug-resistant Virus Strains
J.MOL.BIOL., 324, 2002
|
|
5N06
 
 | |
1IZH
 
 | | Inhibitor of HIV protease with unusual binding mode potently inhibiting multi-resistant protease mutants | | Descriptor: | proteinase, {(1S)-1-BENZYL-4-[3-CARBAMOYL-1-(1-CARBAMOYL-2-PHENYL-ETHYLCARBAMOYL)-(S)-PROPYLCARBAMOYL]-2-OXO-5-PHENYL-PENTYL}-CARBAMIC ACID TERT-BUTYL ESTER | | Authors: | Weber, J, Mesters, J.R, Lepsik, M, Prejdova, J, Svec, M, Sponarova, J, Mlcochova, P, Skalicka, K, Strisovsky, K, Uhlikova, T, Soucek, M, Machala, L, Stankova, M, Vondrasek, J, Klimkait, T, Kraeusslich, H.-G, Hilgenfeld, R, Konvalinka, J. | | Deposit date: | 2002-10-02 | | Release date: | 2002-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unusual Binding Mode of an HIV-1 Protease Inhibitor Explains its Potency against Multi-drug-resistant Virus Strains
J.MOL.BIOL., 324, 2002
|
|
3PUP
 
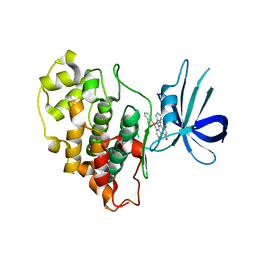 | | Structure of Glycogen Synthase Kinase 3 beta (GSK3B) in complex with a ruthenium octasporine ligand (OS1) | | Descriptor: | Glycogen synthase kinase-3 beta, Ruthenium octasporine | | Authors: | Filippakopoulos, P, Kraling, K, Essen, L.O, Meggers, E, Knapp, S. | | Deposit date: | 2010-12-06 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structurally sophisticated octahedral metal complexes as highly selective protein kinase inhibitors.
J.Am.Chem.Soc., 133, 2011
|
|
1HZ8
 
 | | SOLUTION STRUCTURE AND BACKBONE DYNAMICS OF A CONCATEMER OF EGF-HOMOLOGY MODULES OF THE HUMAN LOW DENSITY LIPOPROTEIN RECEPTOR | | Descriptor: | CALCIUM ION, LOW DENSITY LIPOPROTEIN RECEPTOR | | Authors: | Kurniawan, N.D, Aliabadizadeh, K, Brereton, I.M, Kroon, P.A, Smith, R. | | Deposit date: | 2001-01-23 | | Release date: | 2001-08-15 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | NMR structure and backbone dynamics of a concatemer of epidermal growth factor homology modules of the human low-density lipoprotein receptor.
J.Mol.Biol., 311, 2001
|
|
1I0U
 
 | | SOLUTION STRUCTURE AND BACKBONE DYNAMICS OF A CONCATEMER OF EGF-HOMOLOGY MODULES OF THE HUMAN LOW DENSITY LIPOPROTEIN RECEPTOR | | Descriptor: | CALCIUM ION, LOW DENSITY LIPOPROTEIN RECEPTOR | | Authors: | Kurniawan, N.D, Aliabadizadeh, K, Brereton, I.M, Kroon, P.A, Smith, R. | | Deposit date: | 2001-01-29 | | Release date: | 2001-08-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR structure and backbone dynamics of a concatemer of epidermal growth factor homology modules of the human low-density lipoprotein receptor.
J.Mol.Biol., 311, 2001
|
|
