6DZ8
 
 | |
7KCW
 
 | | Crystal structure of S. aureus penicillin-binding protein 4 (PBP4) mutant (R200L) in complex with nafcillin | | 分子名称: | (2R,4S)-2-[(1R)-1-{[(2-ethoxynaphthalen-1-yl)carbonyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4, ... | | 著者 | Alexander, J.A, Strynadka, N.C. | | 登録日 | 2020-10-07 | | 公開日 | 2021-06-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | PBP4-mediated beta-lactam resistance among clinical strains of Staphylococcus aureus.
J.Antimicrob.Chemother., 76, 2021
|
|
7KCX
 
 | | Crystal structure of S. aureus penicillin-binding protein 4 (PBP4) mutant (R200L) in complex with cefoxitin | | 分子名称: | (2R)-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4, ... | | 著者 | Alexander, J.A.N, Strynadka, N.C.J. | | 登録日 | 2020-10-07 | | 公開日 | 2021-06-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | PBP4-mediated beta-lactam resistance among clinical strains of Staphylococcus aureus.
J.Antimicrob.Chemother., 76, 2021
|
|
7KCV
 
 | |
7KCY
 
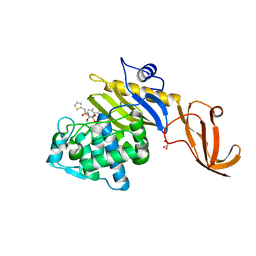 | | Crystal structure of S. aureus penicillin-binding protein 4 (PBP4) with cefoxitin | | 分子名称: | (2R)-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4, ... | | 著者 | Alexander, J.A.N, Strynadka, N.C.J. | | 登録日 | 2020-10-07 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | PBP4-mediated beta-lactam resistance among clinical strains of Staphylococcus aureus.
J.Antimicrob.Chemother., 76, 2021
|
|
6C39
 
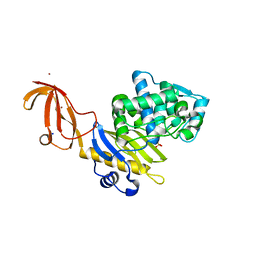 | |
6C3K
 
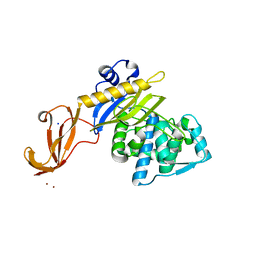 | |
6O9S
 
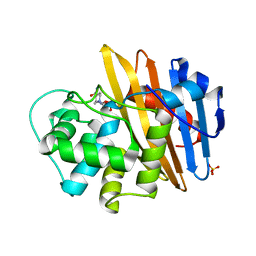 | |
6O9W
 
 | |
8EXS
 
 | | Cryo-EM structure of S. aureus BlaR1 F284A mutant | | 分子名称: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | 著者 | Alexander, J.A.N, Hu, J, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2022-10-25 | | 公開日 | 2023-01-11 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
8EXT
 
 | | Cryo-EM structure of S. aureus BlaR1 F284A mutant in complex with ampicillin | | 分子名称: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | 著者 | Alexander, J.A.N, Hu, J, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2022-10-25 | | 公開日 | 2023-01-11 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
5TW4
 
 | |
5TX9
 
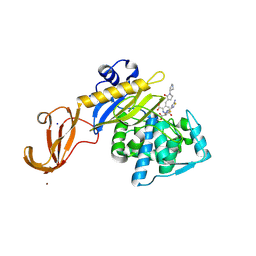 | | Crystal structure of S. aureus penicillin binding protein 4 (PBP4) mutant (E183A, F241R) in complex with ceftobiprole | | 分子名称: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Penicillin-binding protein 4, SODIUM ION, ... | | 著者 | Alexander, J.A.N, Strynadka, N.C.J. | | 登録日 | 2016-11-16 | | 公開日 | 2018-05-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Structural and kinetic analysis of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance inStaphylococcus aureus.
J. Biol. Chem., 2018
|
|
5TW8
 
 | |
5TXI
 
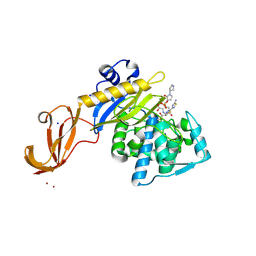 | | Crystal structure of wild-type S. aureus penicillin binding protein 4 (PBP4) in complex with ceftobiprole | | 分子名称: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | 著者 | Alexander, J.A.N, Strynadka, N.C.J. | | 登録日 | 2016-11-16 | | 公開日 | 2018-05-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural and kinetic analysis of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance inStaphylococcus aureus.
J. Biol. Chem., 2018
|
|
5TY2
 
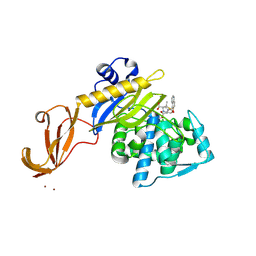 | | Crystal structure of S. aureus penicillin binding protein 4 (PBP4) mutant (E183A, F241R) in complex with nafcillin | | 分子名称: | (2R,4S)-2-[(1R)-1-{[(2-ethoxynaphthalen-1-yl)carbonyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, Penicillin-binding protein 4, ... | | 著者 | Alexander, J.A.N, Strynadka, N.C.J. | | 登録日 | 2016-11-18 | | 公開日 | 2018-06-13 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and kinetic analysis of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance inStaphylococcus aureus.
J. Biol. Chem., 2018
|
|
5TY7
 
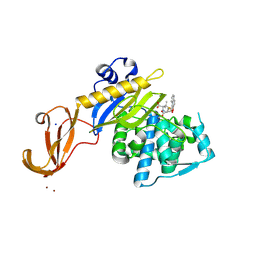 | | Crystal structure of wild-type S. aureus penicillin binding protein 4 (PBP4) in complex with nafcillin | | 分子名称: | (2R,4S)-2-[(1R)-1-{[(2-ethoxynaphthalen-1-yl)carbonyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 4, SODIUM ION, ... | | 著者 | Alexander, J.A.N, Strynadka, N.C.J. | | 登録日 | 2016-11-18 | | 公開日 | 2018-06-13 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.894 Å) | | 主引用文献 | Structural and kinetic analysis of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance inStaphylococcus aureus.
J. Biol. Chem., 2018
|
|
6XFU
 
 | |
8EXP
 
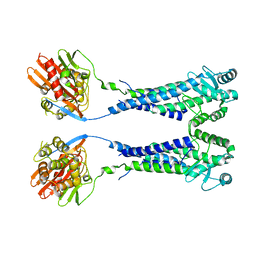 | | Cryo-EM structure of S. aureus BlaR1 with C2 symmetry | | 分子名称: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | 著者 | Worrall, L.J, Alexander, J.A.N, Vuckovic, M, Strynadka, N.C.J. | | 登録日 | 2022-10-25 | | 公開日 | 2023-01-11 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
8EXR
 
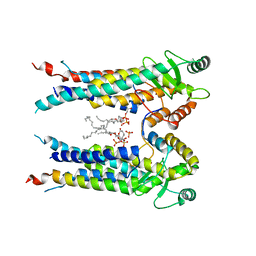 | | Cryo-EM structure of S. aureus BlaR1 TM and zinc metalloprotease domain | | 分子名称: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Beta-lactam sensor/signal transducer BlaR1, PHOSPHATE ION, ... | | 著者 | Worrall, L.J, Alexander, J.A.N, Vuckovic, M, Strynadka, N.C.J. | | 登録日 | 2022-10-25 | | 公開日 | 2023-01-11 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
8EXQ
 
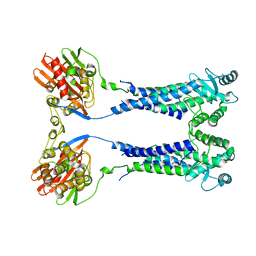 | | Cryo-EM structure of S. aureus BlaR1 with C1 symmetry | | 分子名称: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | 著者 | Worrall, L.J, Alexander, J.A.N, Vuckovic, M, Strynadka, N.C.J. | | 登録日 | 2022-10-25 | | 公開日 | 2023-01-11 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
