2QLJ
 
 | | Crystal Structure of Caspase-7 with Inhibitor AC-WEHD-CHO | | 分子名称: | CITRIC ACID, Caspase-7, Inhibitor AC-WEHD-CHO, ... | | 著者 | Agniswamy, J, Fang, B, Weber, I. | | 登録日 | 2007-07-12 | | 公開日 | 2007-08-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Plasticity of S2-S4 specificity pockets of executioner caspase-7 revealed by structural and kinetic analysis.
Febs J., 274, 2007
|
|
2QL5
 
 | | Crystal Structure of caspase-7 with inhibitor AC-DMQD-CHO | | 分子名称: | CITRIC ACID, Caspase-7, inhibitor, ... | | 著者 | Agniswamy, J, Fang, B, Weber, I. | | 登録日 | 2007-07-12 | | 公開日 | 2007-08-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Plasticity of S2-S4 specificity pockets of executioner caspase-7 revealed by structural and kinetic analysis.
Febs J., 274, 2007
|
|
2QL9
 
 | | Crystal Structure of Caspase-7 with inhibitor AC-DQMD-CHO | | 分子名称: | CITRIC ACID, Caspase-7, Inhibitor AC-DQMD-CHO, ... | | 著者 | Agniswamy, J, Fang, B, Weber, I. | | 登録日 | 2007-07-12 | | 公開日 | 2007-08-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Plasticity of S2-S4 specificity pockets of executioner caspase-7 revealed by structural and kinetic analysis.
Febs J., 274, 2007
|
|
2QLF
 
 | |
5ESC
 
 | |
2QL7
 
 | | Crystal Structure of Caspase-7 with inhibitor AC-IEPD-CHO | | 分子名称: | CITRIC ACID, Caspase-7, Inhibitor AC-IEPD_CHO, ... | | 著者 | Agniswamy, J, Fang, B, Weber, I. | | 登録日 | 2007-07-12 | | 公開日 | 2007-08-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Plasticity of S2-S4 specificity pockets of executioner caspase-7 revealed by structural and kinetic analysis.
Febs J., 274, 2007
|
|
2QLB
 
 | | Crystal Structure of caspase-7 with inhibitor AC-ESMD-CHO | | 分子名称: | CITRIC ACID, Caspase-7, Inhibitor AC-ESMD-CHO, ... | | 著者 | Agniswamy, J, Fang, B, Weber, I. | | 登録日 | 2007-07-12 | | 公開日 | 2007-08-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Plasticity of S2-S4 specificity pockets of executioner caspase-7 revealed by structural and kinetic analysis.
Febs J., 274, 2007
|
|
7MYP
 
 | | Crystal Structure of HIV-1 PRS17 with GRL-44-10A | | 分子名称: | (3R,3aS,4R,6aR)-4-methoxyhexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, GLYCEROL, ... | | 著者 | Agniswamy, J, Weber, I.T. | | 登録日 | 2021-05-21 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Novel HIV PR inhibitors with C4-substituted bis-THF and bis-fluoro-benzyl target the two active site mutations of highly drug resistant mutant PR S17 .
Biochem.Biophys.Res.Commun., 566, 2021
|
|
7MYY
 
 | | Crystal Structure of HIV-1 PRS17 with GRL-142 | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, CHLORIDE ION, GLYCEROL, ... | | 著者 | Agniswamy, J, Weber, I.T. | | 登録日 | 2021-05-22 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Novel HIV PR inhibitors with C4-substituted bis-THF and bis-fluoro-benzyl target the two active site mutations of highly drug resistant mutant PR S17 .
Biochem.Biophys.Res.Commun., 566, 2021
|
|
2AU1
 
 | | Crystal Structure of group A Streptococcus MAC-1 orthorhombic form | | 分子名称: | BETA-MERCAPTOETHANOL, IgG-degrading protease | | 著者 | Agniswamy, J, Nagiec, M.J, Liu, M, Schuck, P, Musser, J.M, Sun, P.D. | | 登録日 | 2005-08-26 | | 公開日 | 2006-02-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of group a streptococcus mac-1: insight into dimer-mediated specificity for recognition of human IgG.
Structure, 14, 2006
|
|
6O5X
 
 | | Crystal Structure of multi-drug resistant HIV-1 protease PR-S17 with substrate analog CA-p2 | | 分子名称: | CHLORIDE ION, HIV-1 protease PR-S17, N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, ... | | 著者 | Agniswamy, J, Wang, Y.-F, Weber, I.T. | | 登録日 | 2019-03-04 | | 公開日 | 2019-06-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Highly Drug-Resistant HIV-1 Protease Mutant PRS17 Shows Enhanced Binding to Substrate Analogues.
Acs Omega, 4, 2019
|
|
2AVW
 
 | | Crystal structure of monoclinic form of streptococcus Mac-1 | | 分子名称: | GLYCEROL, IgG-degrading protease, SULFATE ION | | 著者 | Agniswamy, J, Nagiec, M.J, Liu, M, Schuck, P, Musser, J.M, Sun, P.D. | | 登録日 | 2005-08-30 | | 公開日 | 2006-02-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of group a streptococcus mac-1: insight into dimer-mediated specificity for recognition of human IgG.
Structure, 14, 2006
|
|
5DGW
 
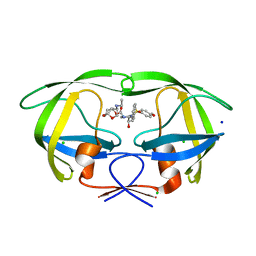 | | Crystal Structure of HIV-1 Protease Inhibitor GRL-105-11A Containing Substituted fused-Tetrahydropyranyl Tetrahydrofuran as P2-Ligand | | 分子名称: | (3R,3aS,4S,7aS)-3-(ethylamino)hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Pol protein, ... | | 著者 | Agniswamy, J, Wang, Y.-F, Weber, I.T. | | 登録日 | 2015-08-28 | | 公開日 | 2015-10-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Design, synthesis, biological evaluation and X-ray structural studies of HIV-1 protease inhibitors containing substituted fused-tetrahydropyranyl tetrahydrofuran as P2-ligands.
Org.Biomol.Chem., 13, 2015
|
|
5DGU
 
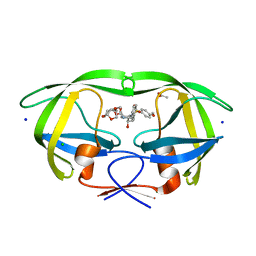 | | Crystal Structure of HIV-1 Protease Inhibitors Containing Substituted fused-Tetrahydropyranyl Tetrahydrofuran as P2-Ligand GRL-004-11A | | 分子名称: | (3R,3aR,4S,7aS)-3-methoxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | 著者 | Agniswamy, J, Wang, Y.-F, Weber, I.T. | | 登録日 | 2015-08-28 | | 公開日 | 2015-10-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Design, synthesis, biological evaluation and X-ray structural studies of HIV-1 protease inhibitors containing substituted fused-tetrahydropyranyl tetrahydrofuran as P2-ligands.
Org.Biomol.Chem., 13, 2015
|
|
5T2E
 
 | |
5T2Z
 
 | | Crystal Structure of Multi-drug Resistant HIV-1 Protease PR-S17 in Complex with Darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | 著者 | Agniswamy, J, Weber, I.T. | | 登録日 | 2016-08-24 | | 公開日 | 2017-01-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural Studies of a Rationally Selected Multi-Drug Resistant HIV-1 Protease Reveal Synergistic Effect of Distal Mutations on Flap Dynamics.
PLoS ONE, 11, 2016
|
|
3EDR
 
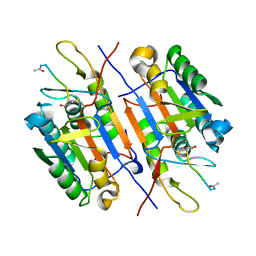 | |
3IBF
 
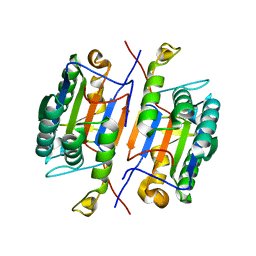 | | Crystal structure of unliganded caspase-7 | | 分子名称: | Caspase-7 | | 著者 | Agniswamy, J. | | 登録日 | 2009-07-15 | | 公開日 | 2009-09-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Conformational similarity in the activation of caspase-3 and -7 revealed by the unliganded and inhibited structures of caspase-7.
Apoptosis, 14, 2009
|
|
3IBC
 
 | |
3TL9
 
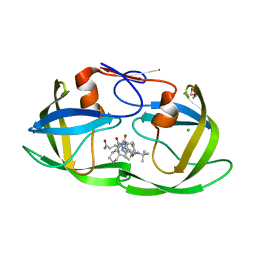 | | crystal structure of HIV protease model precursor/Saquinavir complex | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, GLYCEROL, ... | | 著者 | Agniswamy, J, Sayer, J, Weber, I, Louis, J. | | 登録日 | 2011-08-29 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Terminal interface conformations modulate dimer stability prior to amino terminal autoprocessing of HIV-1 protease.
Biochemistry, 51, 2012
|
|
4YE3
 
 | | Crystal Structure of Multidrug Resistant HIV-1 Protease Clinical Isolate PR20 with Inhibitor GRL-4410A | | 分子名称: | (3R,3aS,4R,6aR)-4-methoxyhexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, GLYCEROL, ... | | 著者 | Agniswamy, J, Weber, I.T. | | 登録日 | 2015-02-23 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Substituted Bis-THF Protease Inhibitors with Improved Potency against Highly Resistant Mature HIV-1 Protease PR20.
J.Med.Chem., 58, 2015
|
|
4YHQ
 
 | | Crystal structure of multidrug resistant clinical isolate PR20 with GRL-5010A | | 分子名称: | (3R,3aS,6aS)-4,4-difluorohexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, GLYCEROL, ... | | 著者 | Agniswamy, J, Weber, I.T. | | 登録日 | 2015-02-27 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Substituted Bis-THF Protease Inhibitors with Improved Potency against Highly Resistant Mature HIV-1 Protease PR20.
J.Med.Chem., 58, 2015
|
|
4Z50
 
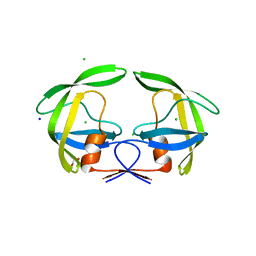 | | Crystal Structure of Multidrug Resistant HIV-1 Protease Clinical Isolate PR20D25N with Tucked Flap | | 分子名称: | CHLORIDE ION, GLYCEROL, Protease, ... | | 著者 | Agniswamy, J, Shen, C.-H, Weber, I.T. | | 登録日 | 2015-04-02 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Conformational variation of an extreme drug resistant mutant of HIV protease.
J.Mol.Graph.Model., 62, 2015
|
|
3TKW
 
 | | Crystal structure of HIV protease model precursor/Darunavir complex | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, GLYCEROL, ... | | 著者 | Agniswamy, J, Sayer, J, Weber, I, Louis, J. | | 登録日 | 2011-08-29 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Terminal interface conformations modulate dimer stability prior to amino terminal autoprocessing of HIV-1 protease.
Biochemistry, 51, 2012
|
|
3TKG
 
 | | crystal structure of HIV model protease precursor/saquinavir complex | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, GLYCEROL, ... | | 著者 | Agniswamy, J, Sayer, J, Weber, I, Louis, J. | | 登録日 | 2011-08-26 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Terminal interface conformations modulate dimer stability prior to amino terminal autoprocessing of HIV-1 protease.
Biochemistry, 51, 2012
|
|
