3KAS
 
 | | Machupo virus GP1 bound to human transferrin receptor 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein, ... | | Authors: | Abraham, J, Corbett, K.D, Harrison, S.C. | | Deposit date: | 2009-10-19 | | Release date: | 2010-03-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for receptor recognition by New World hemorrhagic fever arenaviruses.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6WRW
 
 | | Crystal structure of computationally designed protein 2DS25.5 in complex with the human Transferrin receptor ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Computationally designed protein 2DS25.5, ... | | Authors: | Abraham, J, Coscia, A, Olal, D, Sahtoe, D.D, Baker, D, Clark, L. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Transferrin receptor targeting by de novo sheet extension.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6WRX
 
 | | Crystal structure of computationally designed protein 2DS25.1 in complex with the human Transferrin receptor ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Abraham, J, Coscia, A, Olal, D, Sahtoe, D.D, Baker, D, Clark, L. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Transferrin receptor targeting by de novo sheet extension.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6WRV
 
 | | Crystal structure of computationally designed protein 3DS18 in complex with the human Transferrin receptor ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Computationally designed protein 3DS18, ... | | Authors: | Abraham, J, Baker, D, Sahtoe, D.D, Coscia, A, Clark, L, Olal, D. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Transferrin receptor targeting by de novo sheet extension.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8UA9
 
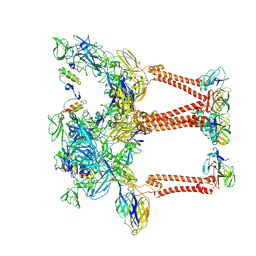 | | Structure of eastern equine encephalitis virus VLP unliganded quasi-threefold spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein, Envelope glycoprotein E1, ... | | Authors: | Abraham, J, Yang, P, Li, W, Fan, X, Pan, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for VLDLR recognition by eastern equine encephalitis virus.
Nat Commun, 15, 2024
|
|
8UA4
 
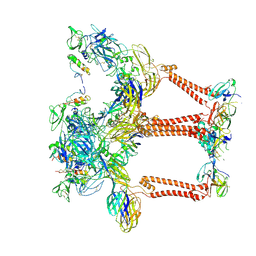 | | Structure of eastern equine encephalitis virus VLP in complex with VLDLR LA1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Capsid protein, ... | | Authors: | Abraham, J, Yang, P, Li, W, Fan, X, Pan, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for VLDLR recognition by eastern equine encephalitis virus.
Nat Commun, 15, 2024
|
|
8UA8
 
 | | Structure of Semliki Forest virus VLP in complex with VLDLR LA2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Assembly protein E3, ... | | Authors: | Abraham, J, Yang, P, Li, W, Fan, X, Pan, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of SFV VLP-VLDLRAD2 complex at the 3-fold axes
To Be Published
|
|
7OXO
 
 | | human LonP1, R-state, incubated in AMPPCP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Abrahams, J.P, Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T. | | Deposit date: | 2021-06-22 | | Release date: | 2021-12-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
1BMF
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE, MAGNESIUM ION, ... | | Authors: | Abrahams, J.P, Leslie, A.G.W, Lutter, R, Walker, J.E. | | Deposit date: | 1996-03-13 | | Release date: | 1996-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria.
Nature, 370, 1994
|
|
1EFR
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE COMPLEXED WITH THE PEPTIDE ANTIBIOTIC EFRAPEPTIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE SUBUNIT ALPHA, BOVINE MITOCHONDRIAL F1-ATPASE SUBUNIT BETA, ... | | Authors: | Abrahams, J.P, Buchanan, S.K, Van Raaij, M.J, Fearnley, I.M, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 1996-05-24 | | Release date: | 1997-02-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Structure of Bovine F1-ATPase Complexed with the Peptide Antibiotic Efrapeptin.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1PSI
 
 | |
1EXS
 
 | | STRUCTURE OF PORCINE BETA-LACTOGLOBULIN | | Descriptor: | BETA-LACTOGLOBULIN, GLYCEROL, SODIUM ION | | Authors: | Abrahams, J.P, Hoedemaeker, F.J. | | Deposit date: | 2000-05-04 | | Release date: | 2000-11-15 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A novel pH-dependent dimerization motif in beta-lactoglobulin from pig (Sus scrofa).
Acta Crystallogr.,Sect.D, 58, 2002
|
|
8EXX
 
 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA and foscarnet (pre-translocation state) | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2022-10-26 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
5EN2
 
 | | Molecular basis for antibody-mediated neutralization of New World hemorrhagic fever mammarenaviruses | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Mahmutovic, S, Clark, L, Levis, S, Briggiler, A, Enria, D, Harrison, S.C, Abraham, J. | | Deposit date: | 2015-11-09 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Molecular Basis for Antibody-Mediated Neutralization of New World Hemorrhagic Fever Mammarenaviruses.
Cell Host Microbe, 18, 2015
|
|
8V1T
 
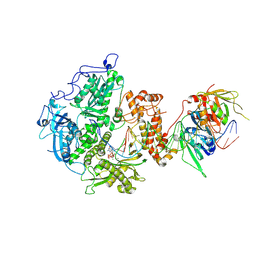 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA and acyclovir triphosphate in closed conformation | | Descriptor: | ACYCLOVIR TRIPHOSPHATE, DNA polymerase, DNA polymerase processivity factor, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
8V1R
 
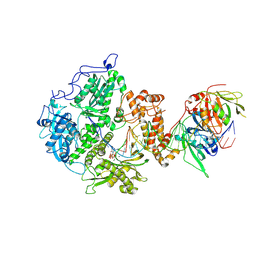 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA and DTTP in closed conformation | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
8V1Q
 
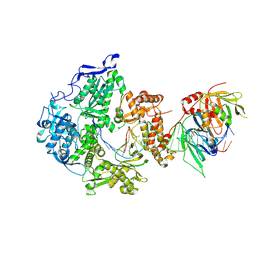 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA in both open/closed conformations | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
8V1S
 
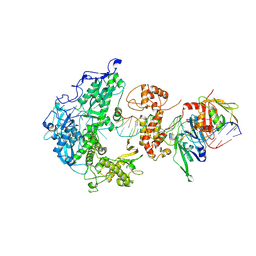 | | Herpes simplex virus 1 polymerase holoenzyme bound to mismatched DNA in editing conformation | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
7SN0
 
 | | Crystal structure of spike protein receptor binding domain of escape mutant SARS-CoV-2 from immunocompromised patient (d146*) in complex with human receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, J, Abraham, J, Clark, S. | | Deposit date: | 2021-10-27 | | Release date: | 2021-12-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain.
Science, 375, 2022
|
|
7SN1
 
 | |
7SN3
 
 | | Structure of human SARS-CoV-2 spike glycoprotein trimer bound by neutralizing antibody C1C-A3 Fab (variable region) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, J, Abraham, J, Shankar, S. | | Deposit date: | 2021-10-27 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain.
Science, 375, 2022
|
|
7SN2
 
 | | Structure of human SARS-CoV-2 neutralizing antibody C1C-A3 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, J, Abraham, J, Yang, P, Shankar, S. | | Deposit date: | 2021-10-27 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain.
Science, 375, 2022
|
|
7T4D
 
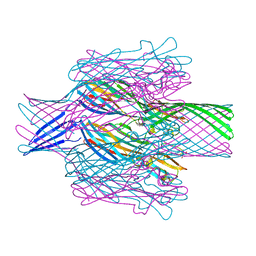 | | Pore structure of pore-forming toxin Epx4 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Epx4 | | Authors: | Xiong, X.Z, Dong, M, Yang, P, Abraham, J. | | Deposit date: | 2021-12-09 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Emerging enterococcus pore-forming toxins with MHC/HLA-I as receptors.
Cell, 185, 2022
|
|
7T4E
 
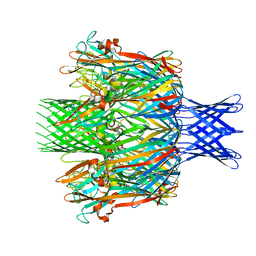 | |
7KFV
 
 | | Structural basis for a germline-biased antibody response to SARS-CoV-2 (RBD:C1A-B12 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of antibody C1A-B12 Fab, Spike glycoprotein, ... | | Authors: | Pan, J, Abraham, J, Clark, L, Clark, S. | | Deposit date: | 2020-10-15 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2.
Biorxiv, 2020
|
|
