2VMI
 
 | | The structure of seleno-methionine labelled CBM51 from Clostridium perfringens GH95 | | Descriptor: | CALCIUM ION, FIBRONECTIN TYPE III DOMAIN PROTEIN | | Authors: | Gregg, K, Finn, R, Abbott, D.W, Boraston, A.B. | | Deposit date: | 2008-01-25 | | Release date: | 2008-02-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Divergent Modes of Glycan Recognition by a New Family of Carbohydrate-Binding Modules
J.Biol.Chem., 283, 2008
|
|
6Y4O
 
 | | Calmodulin bound to cardiac ryanodine receptor (RyR2) calmodulin binding domain | | Descriptor: | CALCIUM ION, Calmodulin-2, Ryanodine receptor 2 | | Authors: | Lau, K, Nielsen, L.H, Holt, C, Brohus, M, Sorensen, A.B, Larsen, K.T, Sommer, C, Van Petegem, F, Overgaard, M.T, Wimmer, R. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83549082 Å) | | Cite: | The arrhythmogenic N53I variant subtly changes the structure and dynamics in the calmodulin N-terminal domain, altering its interaction with the cardiac ryanodine receptor.
J.Biol.Chem., 295, 2020
|
|
2XZ5
 
 | | MMTS-modified Y53C mutant of Aplysia AChBP in complex with acetylcholine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE, ... | | Authors: | Brams, M, Gay, E.A, Colon Saez, J, Guskov, A, van Elk, R, van der Schors, R.C, Peigneur, S, Tytgat, J, Strelkov, S.V, Smit, A.B, Yakel, J.L, Ulens, C. | | Deposit date: | 2010-11-23 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of a Cysteine-Modified Mutant in Loop D of Acetylcholine Binding Protein
J.Biol.Chem., 286, 2011
|
|
6R2W
 
 | | Crystal structure of the super-active FVIIa variant VYT in complex with tissue factor | | Descriptor: | CALCIUM ION, Coagulation factor VII, N-acetyl-D-phenylalanyl-N-[(2S,3S)-6-carbamimidamido-1-chloro-2-hydroxyhexan-3-yl]-L-phenylalaninamide, ... | | Authors: | Sorensen, A.B, Svensson, L.A, Gandhi, P.S. | | Deposit date: | 2019-03-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Beating tissue factor at its own game: Design and properties of a soluble tissue factor-independent coagulation factor VIIa.
J.Biol.Chem., 295, 2020
|
|
6HOJ
 
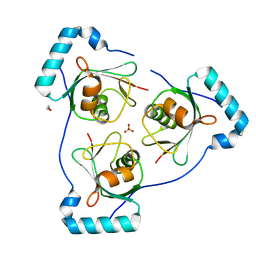 | | Structure of Beclin1 LIR motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, Beclin-1,Gamma-aminobutyric acid receptor-associated protein, SULFATE ION | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
2VIH
 
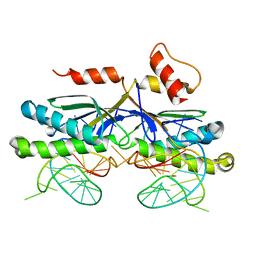 | | CRYSTAL STRUCTURE OF THE IS608 TRANSPOSASE IN COMPLEX WITH Left END 26-MER DNA | | Descriptor: | 5'-D(*AP*AP*AP*GP*CP*CP*CP*CP*TP*AP *GP*CP*TP*TP*TP*TP*AP*GP*CP*TP*AP*TP*GP*GP*GP*G)-3', TRANSPOSASE ORFA | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-12-04 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
2V5N
 
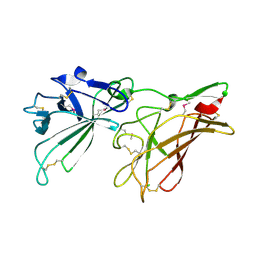 | | STRUCTURE OF HUMAN IGF2R DOMAINS 11-12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR | | Authors: | Brown, J, Delaine, C, Zaccheo, O.J, Siebold, C, Gilbert, R.J, van Boxel, G, Denley, A, Wallace, J.C, Hassan, A.B, Forbes, B.E, Jones, E.Y. | | Deposit date: | 2007-07-06 | | Release date: | 2007-12-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Functional Analysis of the Igf-II/Igf2R Interaction
Embo J., 27, 2008
|
|
6DWW
 
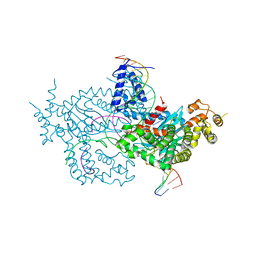 | | Hermes transposase deletion dimer complex with (A/T) DNA and Mn2+ | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*GP*AP*GP*AP*AP*CP*AP*AP*CP*AP*AP*CP*AP*AP*G)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*AP*A)-3'), ... | | Authors: | Dyda, F, Voth, A.R, Hickman, A.B. | | Deposit date: | 2018-06-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Structural insights into the mechanism of double strand break formation by Hermes, a hAT family eukaryotic DNA transposase.
Nucleic Acids Res., 46, 2018
|
|
6Y95
 
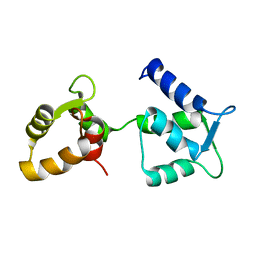 | | Ca2+-free Calmodulin mutant N53I | | Descriptor: | Calmodulin | | Authors: | Holt, C, Hamborg, L.N, Lau, K, Brohus, M, Sorensen, A.B, Larsen, K.T, Sommer, C, Petegem, F.V, Overgaard, M.T, Wimmer, R. | | Deposit date: | 2020-03-06 | | Release date: | 2020-04-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The arrhythmogenic N53I variant subtly changes the structure and dynamics in the calmodulin N-terminal domain, altering its interaction with the cardiac ryanodine receptor.
J.Biol.Chem., 295, 2020
|
|
6WNT
 
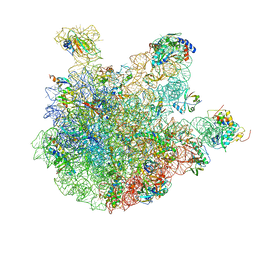 | | 50S ribosomal subunit without free 5S rRNA and perturbed PTC | | Descriptor: | 23s-5s joint ribosomal RNA, 50S ribosomal protein L1, 50S ribosomal protein L10, ... | | Authors: | Loveland, A.B, Korostelev, A.A, Mankin, A.S, Huang, S, Aleksashin, N.A, Klepacki, D, Reier, K, Kefi, A, Szal, A, Remme, J, Jaeger, L, Vazquez-Laslop, N. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ribosome engineering reveals the importance of 5S rRNA autonomy for ribosome assembly.
Nat Commun, 11, 2020
|
|
6DX0
 
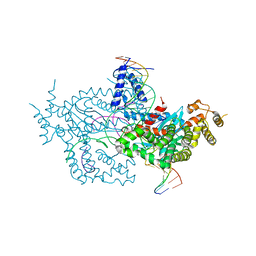 | | Hermes transposase deletion dimer complex with (A/T) DNA | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*GP*AP*GP*AP*AP*CP*AP*AP*CP*AP*AP*CP*AP*AP*G)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*AP*A)-3'), ... | | Authors: | Dyda, F, Voth, A.R, Hickman, A.B. | | Deposit date: | 2018-06-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the mechanism of double strand break formation by Hermes, a hAT family eukaryotic DNA transposase.
Nucleic Acids Res., 46, 2018
|
|
6DLH
 
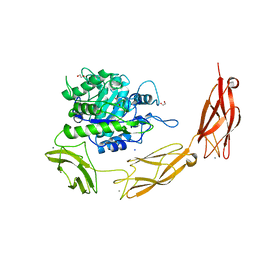 | | Endo-fucoidan hydrolase MfFcnA4 from glycoside hydrolase family 107 | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-endofucoidanase, CALCIUM ION, ... | | Authors: | Vickers, C, Abe, K, Salama-Alber, O, Boraston, A.B. | | Deposit date: | 2018-06-01 | | Release date: | 2018-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to alpha-l-fucosidases from GH29.
J. Biol. Chem., 293, 2018
|
|
6WD5
 
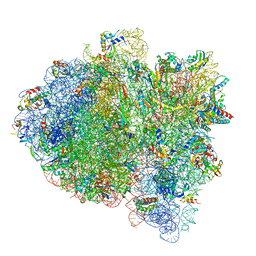 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure II-C1) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
6WDM
 
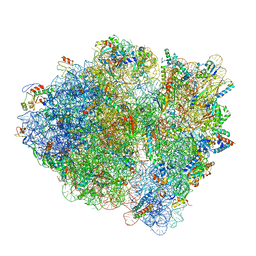 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Non-cognate Structure V-B2) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
6WDD
 
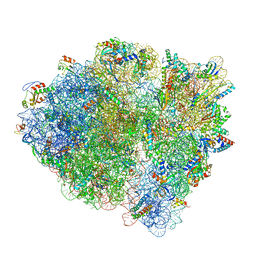 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure V-A) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
2XZ6
 
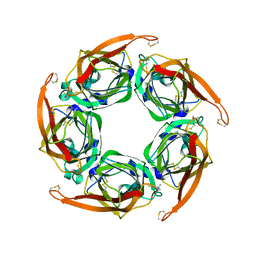 | | MTSET-modified Y53C mutant of Aplysia AChBP | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Brams, M, Gay, E.A, Colon Saez, J, Guskov, A, Van Elk, R, Van Der Schors, R.C, Peigneur, S, Tytgat, J, Strelkov, S.V, Smit, A.B, Yakel, J.L, Ulens, C. | | Deposit date: | 2010-11-23 | | Release date: | 2010-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.137 Å) | | Cite: | Crystal Structures of a Cysteine-Modified Mutant in Loop D of Acetylcholine Binding Protein
J.Biol.Chem., 286, 2011
|
|
6WNW
 
 | | Active 70S ribosome without free 5S rRNA and bound with A- and P- tRNA | | Descriptor: | 16S ribosomal RNA, 23s-5s joint ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Korostelev, A.A, Mankin, A.S, Huang, S, Aleksashin, N.A, Klepacki, D, Reier, K, Kefi, A, Szal, A, Remme, J, Jaeger, L, Vazquez-Laslop, N. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ribosome engineering reveals the importance of 5S rRNA autonomy for ribosome assembly.
Nat Commun, 11, 2020
|
|
2XMA
 
 | | DEINOCOCCUS RADIODURANS ISDRA2 TRANSPOSASE RIGHT END DNA COMPLEX | | Descriptor: | DRA2 TRANSPOSASE RIGHT END RECOGNITION SITE, MAGNESIUM ION, TRANSPOSASE | | Authors: | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | Deposit date: | 2010-07-26 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
6WDF
 
 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure VI-A) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
6WF6
 
 | | Streptomyces coelicolor methylmalonyl-CoA epimerase | | Descriptor: | COBALT (II) ION, DI(HYDROXYETHYL)ETHER, Methylmalonyl-CoA epimerase | | Authors: | Stunkard, L.M, Benjamin, A.B, Bower, J.B, Huth, T.J, Lohman, J.R. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Substrate Enolate Intermediate and Mimic Captured in the Active Site of Streptomyces coelicolor Methylmalonyl-CoA Epimerase.
Chembiochem, 23, 2022
|
|
6WFH
 
 | | Streptomyces coelicolor methylmalonyl-CoA epimerase substrate complex | | Descriptor: | (3S,5R,9R,19E)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9,19-tetrahydroxy-8,8,20-trimethyl-10,14-dioxo-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphahenicos-19-en-21-oic acid 3,5-dioxide (non-preferred name), CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Stunkard, L.M, Benjamin, A.B, Bower, J.B, Huth, T.J, Lohman, J.R. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Substrate Enolate Intermediate and Mimic Captured in the Active Site of Streptomyces coelicolor Methylmalonyl-CoA Epimerase.
Chembiochem, 23, 2022
|
|
2V5O
 
 | | STRUCTURE OF HUMAN IGF2R DOMAINS 11-14 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR, CHLORIDE ION | | Authors: | Brown, J, Delaine, C, Zaccheo, O.J, Siebold, C, Gilbert, R.J, van Boxel, G, Denley, A, Wallace, J.C, Hassan, A.B, Forbes, B.E, Jones, E.Y. | | Deposit date: | 2007-07-06 | | Release date: | 2007-12-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structure and Functional Analysis of the Igf-II/Igf2R Interaction
Embo J., 27, 2008
|
|
2VCB
 
 | | Family 89 Glycoside Hydrolase from Clostridium perfringens in complex with PUGNAc | | Descriptor: | ALPHA-N-ACETYLGLUCOSAMINIDASE, CALCIUM ION, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE | | Authors: | Ficko-Blean, E, Stubbs, K.A, Berg, O, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2007-09-19 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Mechanistic Insight Into the Basis of Mucopolysaccharidosis Iiib.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2VR8
 
 | | Crystal Structure of G85R ALS mutant of Human Cu,Zn Superoxide Dismutase (CuZnSOD) at 1.36 A resolution | | Descriptor: | COPPER (II) ION, SULFATE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | Authors: | Antonyuk, S, Cao, X, Seetharaman, S.V, Whitson, L.J, Taylor, A.B, Holloway, S.P, Strange, R.W, Doucette, P.A, Tiwari, A, Hayward, L.J, Padua, S, Cohlberg, J.A, Selverstone Valentine, J, Hasnain, S.S, Hart, P.J. | | Deposit date: | 2008-03-28 | | Release date: | 2008-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structures of the G85R Variant of Sod1 in Familial Amyotrophic Lateral Sclerosis.
J.Biol.Chem., 283, 2008
|
|
2VNG
 
 | | Family 51 carbohydrate binding module from a family 98 glycoside hydrolase produced by Clostridium perfringens in complex with blood group A-trisaccharide ligand. | | Descriptor: | CALCIUM ION, CPE0329, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Gregg, K.J, Finn, R, Abbott, D.W, Boraston, A.B. | | Deposit date: | 2008-02-04 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Divergent Modes of Glycan Recognition by a New Family of Carbohydrate-Binding Modules
J.Biol.Chem., 283, 2008
|
|
