8GJE
 
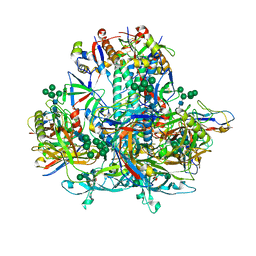 | | HIV-1 Env subtype C CZA97.12 SOSIP.664 in complex with 3BNC117 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3BNC117 Fab heavy chain, ... | | Authors: | Ozorowski, G, Lee, J.H, Ward, A.B. | | Deposit date: | 2023-03-15 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Glycan heterogeneity as a cause of the persistent fraction in HIV-1 neutralization.
Plos Pathog., 19, 2023
|
|
4BMQ
 
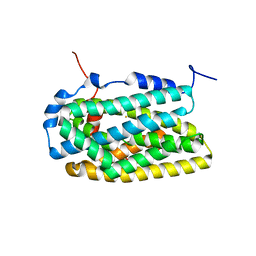 | | Crystal Structure of Ribonucleotide Reductase apo-NrdF from Bacillus cereus (space group C2) | | Descriptor: | FE (II) ION, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE SUBUNIT BETA | | Authors: | Tomter, A.B, Hersleth, H.-P, Hammerstad, M, Rohr, A.K, Andersson, K.K. | | Deposit date: | 2013-05-10 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Bacillus Cereus Class Ib Ribonucleotide Reductase Di-Iron Nrdf in Complex with Nrdi.
Acs Chem.Biol., 9, 2014
|
|
4BQ3
 
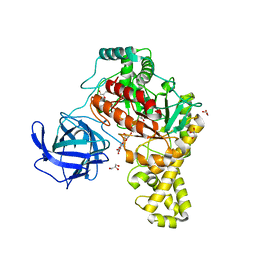 | | Structural analysis of an exo-beta-agarase | | Descriptor: | B-AGARASE, CALCIUM ION, GLYCEROL, ... | | Authors: | Pluvinage, B, Hehemann, J.H, Boraston, A.B. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate Recognition and Hydrolysis by a Family 50 Exo-Beta-Agarase Aga50D from the Marine Bacterium Saccharophagus Degradans
J.Biol.Chem., 288, 2013
|
|
8DC0
 
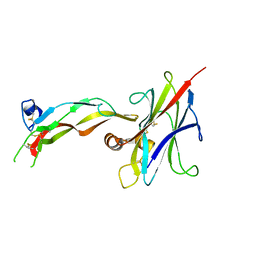 | |
4C21
 
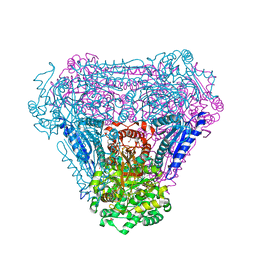 | | L-Fucose Isomerase In Complex With Fucitol | | Descriptor: | 1,2-ETHANEDIOL, FUCITOL, L-FUCOSE ISOMERASE, ... | | Authors: | Higgins, M.A, Suits, M.D.L, Marsters, C, Boraston, A.B. | | Deposit date: | 2013-08-16 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Functional Analysis of Fucose-Processing Enzymes from Streptococcus Pneumoniae.
J.Mol.Biol., 426, 2014
|
|
4BMT
 
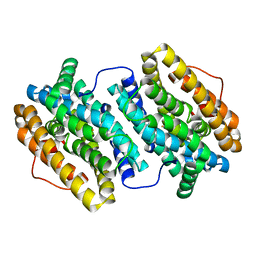 | | Crystal Structure of Ribonucleotide Reductase di-iron NrdF from Bacillus cereus | | Descriptor: | FE (II) ION, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE SUBUNIT BETA | | Authors: | Hersleth, H.-P, Tomter, A.B, Hammerstad, M, Rohr, A.K, Andersson, K.K. | | Deposit date: | 2013-05-10 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Bacillus Cereus Class Ib Ribonucleotide Reductase Di-Iron Nrdf in Complex with Nrdi.
Acs Chem.Biol., 9, 2014
|
|
4C23
 
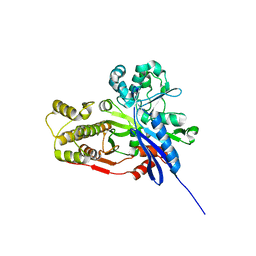 | | L-fuculose kinase | | Descriptor: | 1,2-ETHANEDIOL, L-FUCULOSE KINASE FUCK | | Authors: | Higgins, M.A, Suits, M.D.L, Marsters, C, Boraston, A.B. | | Deposit date: | 2013-08-16 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Analysis of Fucose-Processing Enzymes from Streptococcus Pneumoniae.
J.Mol.Biol., 426, 2014
|
|
4BMU
 
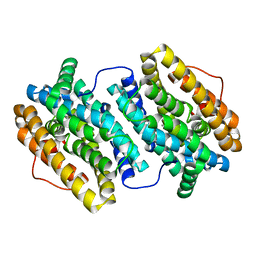 | | Crystal Structure of Ribonucleotide Reductase di-manganese(II) NrdF from Bacillus cereus | | Descriptor: | MANGANESE (II) ION, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE SUBUNIT BETA | | Authors: | Hersleth, H.-P, Tomter, A.B, Hammerstad, M, Rohr, A.K, Andersson, K.K. | | Deposit date: | 2013-05-10 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Bacillus Cereus Class Ib Ribonucleotide Reductase Di-Iron Nrdf in Complex with Nrdi.
Acs Chem.Biol., 9, 2014
|
|
4C24
 
 | | L-fuculose 1-phosphate aldolase | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, L-FUCULOSE PHOSPHATE ALDOLASE, ... | | Authors: | Higgins, M.A, Suits, M.D.L, Marsters, C, Boraston, A.B. | | Deposit date: | 2013-08-16 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Analysis of Fucose-Processing Enzymes from Streptococcus Pneumoniae.
J.Mol.Biol., 426, 2014
|
|
4AWD
 
 | | Crystal structure of the beta-porphyranase BpGH16B (BACPLE_01689) from the human gut bacterium Bacteroides plebeius | | Descriptor: | BETA-PORPHYRANASE, CALCIUM ION, GLYCEROL | | Authors: | Hehemann, J.H, Kelly, A.G, Pudlo, N.A, Martens, E.C, Boraston, A.B. | | Deposit date: | 2012-06-01 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes.
Proc. Natl. Acad. Sci. U.S.A., 109, 2012
|
|
4CC5
 
 | | Fragment-Based Discovery of 6 Azaindazoles As Inhibitors of Bacterial DNA Ligase | | Descriptor: | 2-chloranyl-6-(1H-1,2,4-triazol-3-yl)pyrazine, DNA LIGASE, SULFATE ION | | Authors: | Howard, S, Amin, N, Benowitz, A.B, Chiarparin, E, Cui, H, Deng, X, Heightman, T.D, Holmes, D.J, Hopkins, A, Huang, J, Jin, Q, Kreatsoulas, C, Martin, A.C.L, Massey, F, McCloskey, L, Mortenson, P.N, Pathuri, P, Tisi, D, Williams, P.A. | | Deposit date: | 2013-10-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-Based Discovery of 6-Azaindazoles as Inhibitors of Bacterial DNA Ligase.
Acs Med.Chem.Lett., 4, 2013
|
|
4C18
 
 | |
8EAQ
 
 | | Structure of the full-length IP3R1 channel determined at high Ca2+ | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, CALCIUM ION, Inositol 1,4,5-trisphosphate receptor type 1, ... | | Authors: | Fan, G, Baker, M.R, Terry, L.E, Arige, V, Chen, M, Seryshev, A.B, Baker, M.L, Ludtke, S.J, Yule, D.I, Serysheva, I.I. | | Deposit date: | 2022-08-29 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Conformational motions and ligand-binding underlying gating and regulation in IP 3 R channel.
Nat Commun, 13, 2022
|
|
8EAR
 
 | | Structure of the full-length IP3R1 channel determined in the presence of Calcium/IP3/ATP | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Fan, G, Baker, M.R, Terry, L.E, Arige, V, Chen, M, Seryshev, A.B, Baker, M.L, Ludtke, S.J, Yule, D.I, Serysheva, I.I. | | Deposit date: | 2022-08-29 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformational motions and ligand-binding underlying gating and regulation in IP 3 R channel.
Nat Commun, 13, 2022
|
|
4AAX
 
 | | CpGH89CBM32-5, from Clostridium perfringens, in complex with N- acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, ALPHA-N-ACETYLGLUCOSAMINIDASE, CALCIUM ION, ... | | Authors: | Ficko-Blean, E, Stuart, C.P, Suits, M.D, Cid, M, Tessier, M, Woods, R.J, Boraston, A.B. | | Deposit date: | 2011-12-05 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Carbohydrate Recognition by an Architecturally Complex Alpha-N-Acetylglucosaminidase from Clostridium Perfringens.
Plos One, 7, 2012
|
|
4A34
 
 | |
4A42
 
 | | CpGH89CBM32-6 produced by Clostridium perfringens | | Descriptor: | ALPHA-N-ACETYLGLUCOSAMINIDASE FAMILY PROTEIN, CALCIUM ION | | Authors: | Ficko-Blean, E, Stuart, C.P, Suits, M.D, Cid, M, Tessier, M, Woods, R.J, Boraston, A.B. | | Deposit date: | 2011-10-06 | | Release date: | 2012-04-04 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Carbohydrate Recognition by an Architecturally Complex Alpha-N-Acetylglucosaminidase from Clostridium Perfringens.
Plos One, 7, 2012
|
|
4BMR
 
 | | Crystal Structure of Ribonucleotide Reductase apo-NrdF from Bacillus cereus (space group P21) | | Descriptor: | FE (II) ION, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE SUBUNIT BETA | | Authors: | Hersleth, H.-P, Tomter, A.B, Hammerstad, M, Rohr, A.K, Andersson, K.K. | | Deposit date: | 2013-05-10 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Bacillus Cereus Class Ib Ribonucleotide Reductase Di-Iron Nrdf in Complex with Nrdi.
Acs Chem.Biol., 9, 2014
|
|
4C1R
 
 | | Bacteroides thetaiotaomicron VPI-5482 mannosyl-6-phosphatase Bt3783 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, MANNOSYL-6-PHOSPHATASE | | Authors: | Cuskin, F, Lowe, E.C, Zhu, Y, Temple, M, Thompson, A.J, Cartmell, A, Piens, K, Bracke, D, Vervecken, W, Munoz-Munoz, J.L, Suits, M.D.L, Boraston, A.B, Williams, S.J, Davies, G.J, Abbott, W.D, Martens, E.C, Gilbert, H.J. | | Deposit date: | 2013-08-13 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human Gut Bacteroidetes Can Utilize Yeast Mannan Through a Selfish Mechanism.
Nature, 517, 2015
|
|
4C20
 
 | | L-Fucose Isomerase | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, L-FUCOSE ISOMERASE, ... | | Authors: | Higgins, M.A, Suits, M.D.L, Marsters, C, Boraston, A.B. | | Deposit date: | 2013-08-16 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural and Functional Analysis of Fucose-Processing Enzymes from Streptococcus Pneumoniae.
J.Mol.Biol., 426, 2014
|
|
8DT8
 
 | | LM18/Nb136 bispecific tetra-nanobody immunoglobulin in complex with SARS-CoV-2-6P-Mut7 S protein (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LM18 nanobody, Nb136 nanobody, ... | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2022-07-25 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Fully synthetic platform to rapidly generate tetravalent bispecific nanobody-based immunoglobulins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4A4A
 
 | | CpGH89 (E483Q, E601Q), from Clostridium perfringens, in complex with its substrate GlcNAc-alpha-1,4-galactose | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-beta-D-galactopyranose, ALPHA-N-ACETYLGLUCOSAMINIDASE FAMILY PROTEIN, CALCIUM ION, ... | | Authors: | Ficko-Blean, E, Stuart, C.P, Suits, M.D, Cid, M, Tessier, M, Woods, R.J, Boraston, A.B. | | Deposit date: | 2011-10-07 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of a bacterial exo-alpha-D-N-acetylglucosaminidase in complex with an unusual disaccharide found in class III mucin.
Glycobiology, 22, 2012
|
|
4A6O
 
 | | CpGH89CBM32-4, produced by Clostridium perfringens, in complex with glcNAc-alpha-1,4-galactose | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-beta-D-galactopyranose, ALPHA-N-ACETYLGLUCOSAMINIDASE FAMILY PROTEIN, CALCIUM ION | | Authors: | Ficko-Blean, E, Stuart, C.P, Suits, M.D, Cid, M, Tessier, M, Woods, R.J, Boraston, A.B. | | Deposit date: | 2011-11-07 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Carbohydrate Recognition by an Architecturally Complex Alpha-N-Acetylglucosaminidase from Clostridium Perfringens.
Plos One, 7, 2012
|
|
8DYT
 
 | | Cryo-EM structure of 227 Fab in complex with (NPNA)8 peptide | | Descriptor: | 227 Fab heavy chain, 227 Fab light chain, Circumsporozoite protein | | Authors: | Martin, G.M, Ward, A.B. | | Deposit date: | 2022-08-05 | | Release date: | 2023-08-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Affinity-matured homotypic interactions induce spectrum of PfCSP structures that influence protection from malaria infection.
Nat Commun, 14, 2023
|
|
8E5R
 
 | | Schistosoma mansoni (Blood Fluke) Sulfotransferase/CIDD-0150610 Complex | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase oxamniquine resistance protein, [4-({[(3R)-1-[(1H-indol-3-yl)methyl]-3-{[4-(trifluoromethyl)phenyl]methyl}pyrrolidin-3-yl]methyl}amino)-3-nitrophenyl]methanol | | Authors: | Taylor, A.B, Alwan, S.N. | | Deposit date: | 2022-08-22 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Oxamniquine derivatives overcome Praziquantel treatment limitations for Schistosomiasis.
Plos Pathog., 19, 2023
|
|
