8C54
 
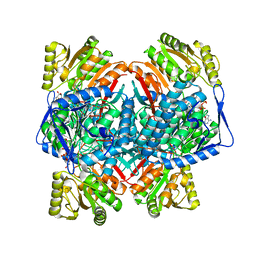 | | Cryo-EM structure of NADH bound SLA dehydrogenase RlGabD from Rhizobium leguminosarum bv. trifolii SRD1565 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Succinate semialdehyde dehydrogenase | | Authors: | Sharma, M, Meek, R.W, Armstrong, Z, Blaza, J.N, Alhifthi, A, Li, J, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2023-01-06 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Molecular basis of sulfolactate synthesis by sulfolactaldehyde dehydrogenase from Rhizobium leguminosarum.
Chem Sci, 14, 2023
|
|
8Q0S
 
 | | X-ray structure of the single chain monellin derivative MNEI | | Descriptor: | ACETATE ION, GLYCEROL, Monellin chain B,Monellin chain A, ... | | Authors: | Ferraro, G, Merlino, A, Lucignano, R, Picone, D. | | Deposit date: | 2023-07-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structural insights and aggregation propensity of a super-stable monellin mutant: A new potential building block for protein-based nanostructured materials.
Int.J.Biol.Macromol., 254, 2024
|
|
3ZBG
 
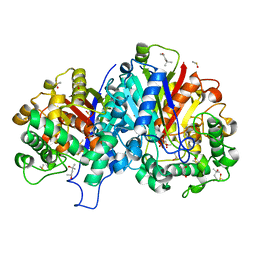 | | Crystal structure of wild-type SCP2 thiolase from Leishmania mexicana at 1.85 A | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-KETOACYL-COA THIOLASE-LIKE PROTEIN, DIMETHYL SULFOXIDE | | Authors: | Harijan, R.K, Kiema, T.-R, Weiss, M.S, Michels, P.A.M, Wierenga, R.K. | | Deposit date: | 2012-11-09 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Scp2-Thiolases of Trypanosomatidae, Human Pathogens Causing Widespread Tropical Diseases: The Importance for Catalysis of the Cysteine of the Unique Hdcf Loop.
Biochem.J., 455, 2013
|
|
7Z3P
 
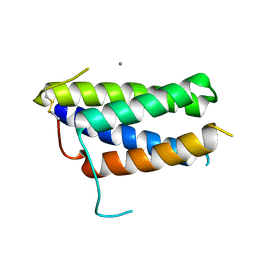 | | Crystal structure of the mouse leptin:LepR-CRH2 encounter complex to 1.95 A resolution. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Leptin, ... | | Authors: | Tsirigotaki, A, Verschueren, K, Savvides, S.N, Verstraete, K. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4MV5
 
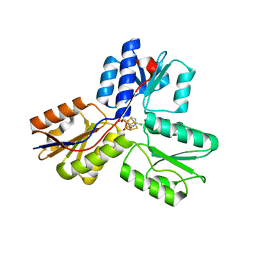 | | IspH in complex with 6-chloropyridin-3-ylmethyl diphosphate | | Descriptor: | (6-chloropyridin-3-yl)methyl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER | | Authors: | Span, I, Wang, K, Song, Y, Eisenreich, W, Bacher, A, Oldfield, E, Groll, M. | | Deposit date: | 2013-09-23 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the Binding of Pyridines to the Iron-Sulfur Enzyme IspH.
J.Am.Chem.Soc., 136, 2014
|
|
7Z4S
 
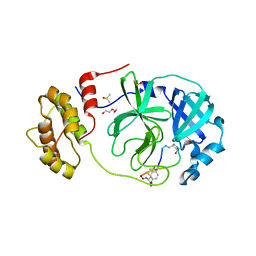 | | Crystal structure of SARS-CoV-2 Mpro in complex with cyclic peptide GM4 including unnatural amino acids. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Owen, C.D, Miura, T, Malla, T, Lukacik, L, Strain-Damerell, C.M, Tumber, A, Brewitz, L, McDonough, M.A, Salah, E, Terasaka, N, Katoh, T, Kawamura, A, Schofield, C.J, Suga, H, Walsh, M.A. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In vitro selection of macrocyclic peptide inhibitors containing cyclic gamma 2,4 -amino acids targeting the SARS-CoV-2 main protease.
Nat.Chem., 15, 2023
|
|
2P5Z
 
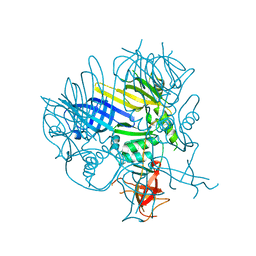 | | The E. coli c3393 protein is a component of the type VI secretion system and exhibits structural similarity to T4 bacteriophage tail proteins gp27 and gp5 | | Descriptor: | Type VI secretion system component | | Authors: | Ramagopal, U.A, Bonanno, J.B, Sridhar, V, Lau, C, Toro, R, Gheyi, T, Maletic, M, Freeman, J.C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin.
Proc.Natl.Acad.Sci.Usa, 106, 2009
|
|
3ZKX
 
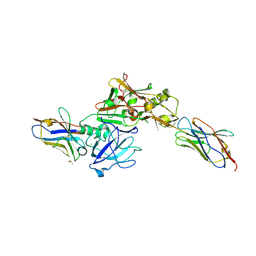 | | TERNARY BACE2 XAPERONE COMPLEX | | Descriptor: | BETA-SECRETASE 2, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Kuglstatter, A, Banner, D.W, Benz, J, Bertschinger, J, Burger, D, Cuppuleri, S, Debulpaep, M, Gast, A, Grabulovski, D, Gsell, B, Hilpert, H, Huber, W, Kusznir, E, Laeremans, T, Matile, H, Rufer, A, Schlatter, D, Steyeart, J, Stihle, M, Thoma, R, Weber, M, Ruf, A. | | Deposit date: | 2013-01-25 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1KWI
 
 | | Crystal Structure Analysis of the Cathelicidin Motif of Protegrins | | Descriptor: | Protegrin-3 Precursor | | Authors: | Sanchez, J.F, Hoh, F, Strub, M.P, Aumelas, A, Dumas, C. | | Deposit date: | 2002-01-29 | | Release date: | 2002-10-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of the cathelicidin motif of protegrin-3 precursor: structural insights into the activation mechanism of an antimicrobial protein.
Structure, 10, 2002
|
|
3ZMV
 
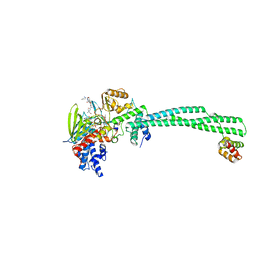 | | LSD1-CoREST in complex with PLSFLV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PKSFLV PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
2PC4
 
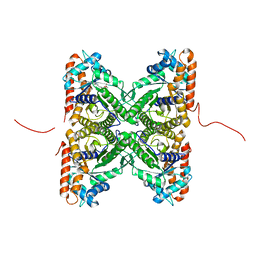 | | Crystal structure of fructose-bisphosphate aldolase from Plasmodium falciparum in complex with TRAP-tail determined at 2.4 angstrom resolution | | Descriptor: | Fructose-bisphosphate aldolase, PbTRAP | | Authors: | Bosch, J, Buscaglia, C.A, Krumm, B, Cardozo, T, Nussenzweig, V, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2007-03-29 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Aldolase provides an unusual binding site for thrombospondin-related anonymous protein in the invasion machinery of the malaria parasite.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3ZPO
 
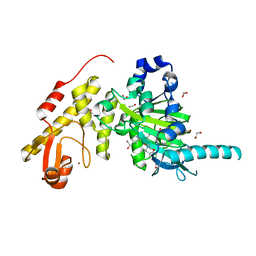 | | Crystal structure of JmjC domain of human histone demethylase UTY with bound GSK J1 | | Descriptor: | 1,2-ETHANEDIOL, 3-[[2-pyridin-2-yl-6-(1,2,4,5-tetrahydro-3-benzazepin-3-yl)pyrimidin-4-yl]amino]propanoic acid, FE (II) ION, ... | | Authors: | Vollmar, M, Gileadi, C, Shrestha, L, Goubin, S, Johansson, C, Krojer, T, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2013-02-28 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human Uty(Kdm6C) is a Male-Specific Nepislon-Methyl Lysyl Demethylase.
J.Biol.Chem., 289, 2014
|
|
6E5S
 
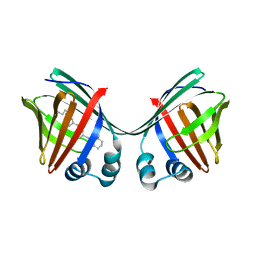 | |
5B1L
 
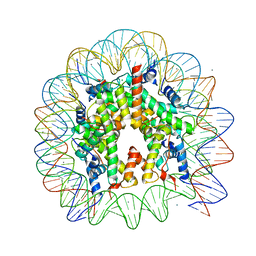 | | The mouse nucleosome structure containing H3t | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1, ... | | Authors: | Urahama, T, Machida, S, Horikoshi, N, Osakabe, A, Tachiwana, H, Taguchi, H, Kurumizaka, H. | | Deposit date: | 2015-12-08 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Testis-Specific Histone Variant H3t Gene Is Essential for Entry into Spermatogenesis
Cell Rep, 18, 2017
|
|
3ZCW
 
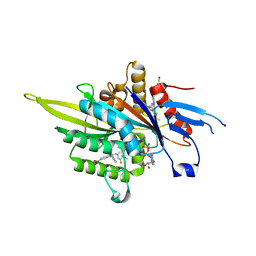 | | Eg5 - New allosteric binding site | | Descriptor: | (2E)-2-(3-fluoranyl-4-methoxy-phenyl)imino-1-[[2-(trifluoromethyl)phenyl]methyl]-3H-benzimidazole-5-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ulaganathan, V, Talapatra, S.K, Kozielski, F, Pannifer, A.D. | | Deposit date: | 2012-11-23 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.691 Å) | | Cite: | Structural Insights Into a Unique Inhibitor Binding Pocket in Kinesin Spindle Protein.
J.Am.Chem.Soc., 135, 2013
|
|
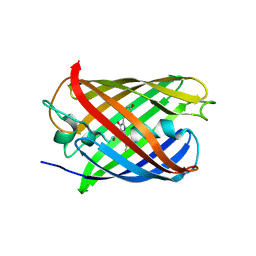
7ZCT
 
 | |
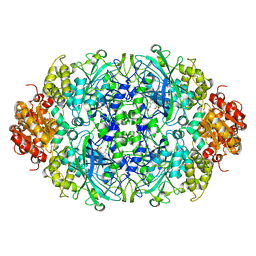
3ZJ4
 
 | |
4N6W
 
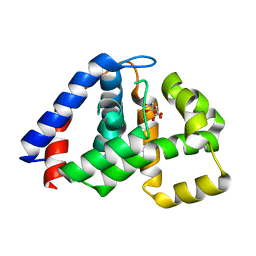 | | X-Ray Crystal Structure of Citrate-bound PhnZ | | Descriptor: | CITRATE ANION, FE (III) ION, Predicted HD phosphohydrolase PhnZ | | Authors: | Worsdorfer, B, Lingaraju, M, Yennawar, N.H, Boal, A.K, Krebs, C, Bollinger Jr, J.M, Pandelia, M.E. | | Deposit date: | 2013-10-14 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Organophosphonate-degrading PhnZ reveals an emerging family of HD domain mixed-valent diiron oxygenases.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6EIG
 
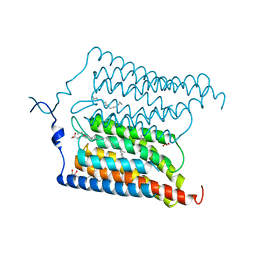 | | Crystal structure of N24Q/C128T mutant of Channelrhodopsin 2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Archaeal-type opsin 2, EICOSANE, ... | | Authors: | Kovalev, K, Borshchevskiy, V, Volkov, O, Polovinkin, V, Marin, E, Balandin, T, Astashkin, R, Bamann, C, Bueldt, G, Willlbold, D, Popov, A, Bamberg, E, Gordeliy, V. | | Deposit date: | 2017-09-19 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into ion conduction by channelrhodopsin 2.
Science, 358, 2017
|
|
1UWR
 
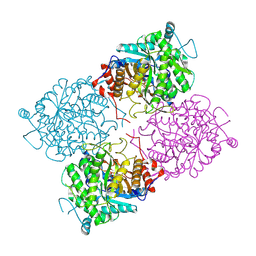 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with 2-deoxy-2-fluoro-galactose | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-galactopyranose, ACETATE ION, BETA-GALACTOSIDASE | | Authors: | Gloster, T.M, Roberts, S, Ducros, V.M.-A, Perugino, G, Rossi, M, Hoos, R, Moracci, M, Vasella, A, Davies, G.J. | | Deposit date: | 2004-02-11 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Studies of the Beta-Glycosidase from Sulfolobus Solfataricus in Complex with Covalently and Noncovalently Bound Inhibitors.
Biochemistry, 43, 2004
|
|
1UJ2
 
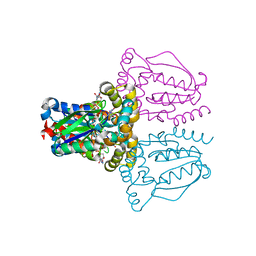 | | Crystal structure of human uridine-cytidine kinase 2 complexed with products, CMP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CYTIDINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Suzuki, N.N, Koizumi, K, Fukushima, M, Matsuda, A, Inagaki, F. | | Deposit date: | 2003-07-25 | | Release date: | 2004-05-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the specificity, catalysis, and regulation of human uridine-cytidine kinase
STRUCTURE, 12, 2004
|
|
4N8M
 
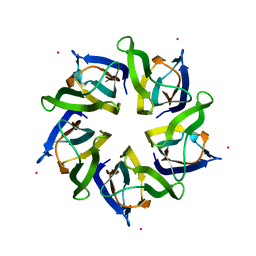 | | Structural polymorphism in the N-terminal oligomerization domain of NPM1 | | Descriptor: | COBALT (II) ION, Nucleophosmin | | Authors: | Mitrea, D, Royappa, G, Buljan, M, Yun, M, Pytel, N, Satumba, J, Nourse, A, Park, C, Babu, M.M, White, S.W, Kriwacki, R.W. | | Deposit date: | 2013-10-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural polymorphism in the N-terminal oligomerization domain of NPM1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3ZN1
 
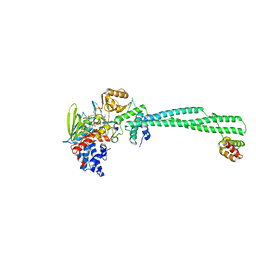 | | LSD1-CoREST in complex with PRLYLV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-13 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
6EKU
 
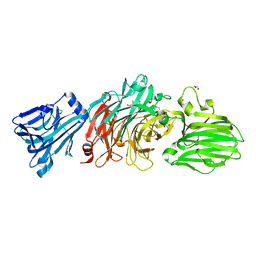 | | Vibrio cholerae neuraminidase complexed with zanamivir | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Schuetz, A, Heinemann, U. | | Deposit date: | 2017-09-27 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Different Inhibitory Potencies of Oseltamivir Carboxylate, Zanamivir, and Several Tannins on Bacterial and Viral Neuraminidases as Assessed in a Cell-Free Fluorescence-Based Enzyme Inhibition Assay.
Molecules, 22, 2017
|
|
3ZBZ
 
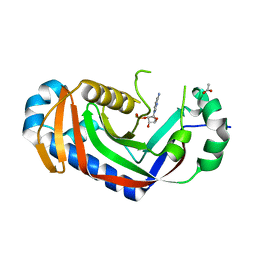 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation V321A, crystallized with 2'-AMPS | | Descriptor: | 2', 3'-CYCLIC-NUCLEOTIDE 3'-PHOSPHODIESTERASE, 2'-O-(sulfidophosphinato)adenosine, ... | | Authors: | Myllykoski, M, Raasakka, A, Lehtimaki, M, Han, H, Kursula, P. | | Deposit date: | 2012-11-14 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic Analysis of the Reaction Cycle of 2',3'-Cyclic Nucleotide 3'-Phosphodiesterase, a Unique Member of the 2H Phosphoesterase Family
J.Mol.Biol., 425, 2013
|
|
