7LRA
 
 | |
6I0T
 
 | | Crystal structure of DmTailor in complex with GpU | | Descriptor: | RNA (5'-R(*GP*U)-3'), Terminal uridylyltransferase Tailor | | Authors: | Kroupova, A, Ivascu, A, Jinek, M. | | Deposit date: | 2018-10-26 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for acceptor RNA substrate selectivity of the 3' terminal uridylyl transferase Tailor.
Nucleic Acids Res., 47, 2019
|
|
6NZP
 
 | |
1CVK
 
 | | T4 LYSOZYME MUTANT L118A | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-23 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
7QG6
 
 | | Co-crystal structure of UPF3A-RRM-NOPS-L with UPF2-MIF4GIII | | Descriptor: | CHLORIDE ION, Regulator of nonsense transcripts 2, Regulator of nonsense transcripts 3A, ... | | Authors: | Powers, K.T, Bufton, J.C, Szeto, J.A, Schaffitzel, C. | | Deposit date: | 2021-12-07 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of nonsense-mediated mRNA decay factors UPF3B and UPF3A in complex with UPF2 reveal molecular basis for competitive binding and for neurodevelopmental disorder-causing mutation.
Nucleic Acids Res., 50, 2022
|
|
7LRV
 
 | |
6O3C
 
 | | Crystal structure of active Smoothened bound to SAG21k, cholesterol, and NbSmo8 | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-chloro-4,7-difluoro-N-{[2-methoxy-5-(pyridin-4-yl)phenyl]methyl}-N-[trans-4-(methylamino)cyclohexyl]-1-benzothiophene-2-carboxamide, ... | | Authors: | Deshpande, I.S, Liang, J, Hedeen, D, Roberts, K.J, Zhang, Y, Ha, B, Latorraca, N.R, Faust, B, Dror, R.O, Beachy, P.A, Myers, B.R, Manglik, A. | | Deposit date: | 2019-02-26 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Smoothened stimulation by membrane sterols drives Hedgehog pathway activity.
Nature, 571, 2019
|
|
6Z2T
 
 | |
6KXF
 
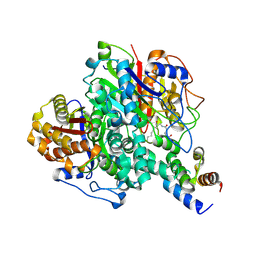 | | The ishigamide ketosynthase/chain length factor | | Descriptor: | ACP, Ketosynthase, [(3~{R})-2,2-dimethyl-4-[[3-[2-[[(~{E})-oct-2-enoyl]amino]ethylamino]-3-oxidanylidene-propyl]amino]-3-oxidanyl-4-oxidanylidene-butyl] dihydrogen phosphate | | Authors: | Du, D, Katsuyama, Y, Horiuchi, M, Fushinobu, S, Chen, A, Davis, T, Burkart, M, Ohnishi, Y. | | Deposit date: | 2019-09-10 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for selectivity in a highly reducing type II polyketide synthase.
Nat.Chem.Biol., 16, 2020
|
|
1EZK
 
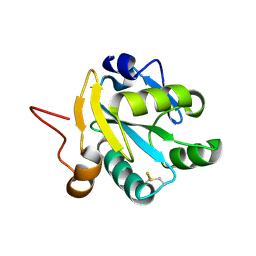 | | Crystal structure of recombinant tryparedoxin I | | Descriptor: | TRYPAREDOXIN I | | Authors: | Hofmann, B, Guerrero, S.A, Kalisz, H.M, Menge, U, Nogoceke, E, Montemartini, M, Singh, M, Flohe, L, Hecht, H.J. | | Deposit date: | 2000-05-11 | | Release date: | 2000-05-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of tryparedoxins revealing interaction with trypanothione.
Biol.Chem., 382, 2001
|
|
5LH9
 
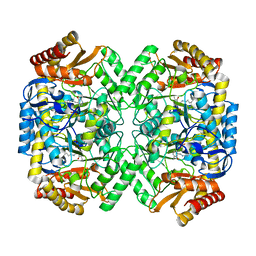 | |
1D4D
 
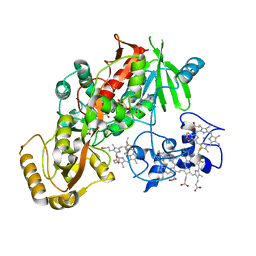 | | CRYSTAL STRUCTURE OF THE SUCCINATE COMPLEXED FORM OF THE FLAVOCYTOCHROME C FUMARATE REDUCTASE OF SHEWANELLA PUTREFACIENS STRAIN MR-1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FLAVOCYTOCHROME C FUMARATE REDUCTASE, HEME C, ... | | Authors: | Leys, D, Tsapin, A.S, Meyer, T.E, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 1999-10-03 | | Release date: | 1999-12-01 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of the flavocytochrome c fumarate reductase of Shewanella putrefaciens MR-1.
Nat.Struct.Biol., 6, 1999
|
|
4XF4
 
 | | Cysteine dioxygenase at pH 8.0 in complex with homocysteine | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Cysteine dioxygenase type 1, FE (III) ION | | Authors: | Driggers, C.M, Karplus, P.A. | | Deposit date: | 2014-12-26 | | Release date: | 2016-03-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3485 Å) | | Cite: | Structure-Based Insights into the Role of the Cys-Tyr Crosslink and Inhibitor Recognition by Mammalian Cysteine Dioxygenase.
J. Mol. Biol., 428, 2016
|
|
8FAT
 
 | | Crystal structure of Ky224 Fab in complex with circumsporozoite protein NPDP peptide | | Descriptor: | Circumsporozoite protein NPDP peptide, Ky224 Antibody, heavy chain, ... | | Authors: | Kassardjian, A, Thai, E, Julien, J.P. | | Deposit date: | 2022-11-28 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular determinants of cross-reactivity and potency by VH3-33 antibodies against the Plasmodium falciparum circumsporozoite protein.
Cell Rep, 42, 2023
|
|
4XFH
 
 | |
6NVZ
 
 | |
5LEG
 
 | | Structure of the bacterial sex F pilus (pED208) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, Pilin | | Authors: | Costa, T.R.D, Ilangovan, I, Ukleja, M, Redzej, A, Santini, J.M, Smith, T.K, Egelman, E.H, Waksman, G. | | Deposit date: | 2016-06-29 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the Bacterial Sex F Pilus Reveals an Assembly of a Stoichiometric Protein-Phospholipid Complex.
Cell, 166, 2016
|
|
8PS2
 
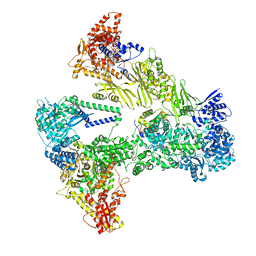 | | Asymmetric unit of the yeast fatty acid synthase with ACP at the enoyl reductase domain (FASam sample) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta, ... | | Authors: | Singh, K, Bunzel, G, Graf, B, Yip, K.M, Stark, H, Chari, A. | | Deposit date: | 2023-07-13 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Cell, 186, 2023
|
|
8PRV
 
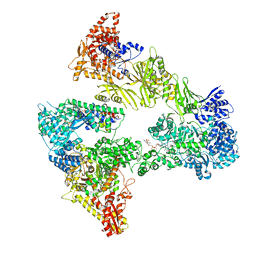 | | Asymmetric unit of the yeast fatty acid synthase in the non-rotated state with ACP at the ketosreductase domain (FASamn sample) | | Descriptor: | COENZYME A, FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, ... | | Authors: | Singh, K, Bunzel, G, Graf, B, Yip, K.M, Stark, H, Chari, A. | | Deposit date: | 2023-07-12 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Cell, 186, 2023
|
|
7LRB
 
 | |
3R8P
 
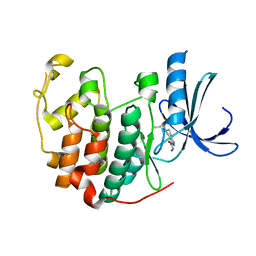 | | CDK2 in complex with inhibitor NSK-MC1-6 | | Descriptor: | (5R)-5-propyl-4,5,6,7-tetrahydro-1H-indazole-3-carbohydrazide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
8CXE
 
 | |
1D8M
 
 | | CRYSTAL STRUCTURE OF MMP3 COMPLEXED WITH A HETEROCYCLE-BASED INHIBITOR | | Descriptor: | 1-BENZYL-3-(4-METHOXY-BENZENESULFONYL)-6-OXO-HEXAHYDRO-PYRIMIDINE-4-CARBOXYLIC ACID HYDROXYAMIDE, CALCIUM ION, STROMELYSIN-1 PRECURSOR, ... | | Authors: | Pikul, S, Dunham, K.M, Almstead, N.G, De, B, Natchus, M.G, Taiwo, Y.O, Williams, L.E, Hynd, B.A, Hsieh, L.C, Janusz, M.J, Gu, F, Mieling, G.E. | | Deposit date: | 1999-10-25 | | Release date: | 2000-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Heterocycle-based MMP inhibitors with P2' substituents.
Bioorg.Med.Chem.Lett., 11, 2001
|
|
8PRW
 
 | | Cryo-EM structure of the yeast fatty acid synthase at 1.9 angstrom resolution | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, COENZYME A, Fatty acid synthase subunit alpha, ... | | Authors: | Singh, K, Bunzel, G, Graf, B, Yip, K.M, Stark, H, Chari, A. | | Deposit date: | 2023-07-12 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Cell, 186, 2023
|
|
6HQA
 
 | | Molecular structure of promoter-bound yeast TFIID | | Descriptor: | Histone-fold, Subunit (17 kDa) of TFIID and SAGA complexes, involved in RNA polymerase II transcription initiation, ... | | Authors: | Kolesnikova, O, Ben-Shem, A, Luo, J, Ranish, J, Schultz, P, Papai, G. | | Deposit date: | 2018-09-24 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Molecular structure of promoter-bound yeast TFIID.
Nat Commun, 9, 2018
|
|
