8FE4
 
 | | Structure of dengue virus (DENV2) in complex with prM13, an anti-PrM monoclonal antibody | | Descriptor: | Envelope protein E, prM protein, prM13 Fab Heavy Chain, ... | | Authors: | Dowd, A.D, Sirohi, D, Speer, S, Mukherjee, S, Govero, J, Aleshnick, M, Larman, B, Sukupolvi-Petty, S, Sevvana, M, Miller, A.S, Klose, T, Zheng, A, Kielian, M, Kuhn, R.J, Diamond, M.S, Pierson, T.C. | | Deposit date: | 2022-12-05 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | prM-reactive antibodies reveal a role for partially mature virions in dengue virus pathogenesis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7Q8Y
 
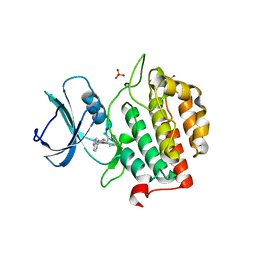 | | Crystal structure of TTBK2 in complex with VNG2.73 (compound 42) | | Descriptor: | PHOSPHATE ION, Tau-tubulin kinase 2, ~{N}-[4-(2-chloranylphenoxy)phenyl]-7~{H}-pyrrolo[2,3-d]pyrimidin-4-amine | | Authors: | Chaikuad, A, Nozal, V, Martinez, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-11 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | TDP-43 Modulation by Tau-Tubulin Kinase 1 Inhibitors: A New Avenue for Future Amyotrophic Lateral Sclerosis Therapy.
J.Med.Chem., 65, 2022
|
|
8FE3
 
 | | Structure of dengue virus (DENV2) in complex with prM12, an anti-PrM monoclonal antibody | | Descriptor: | Envelope protein E, prM protein, prM12 Fab Heavy Chain, ... | | Authors: | Dowd, A.D, Sirohi, D, Speer, S, Mukherjee, S, Govero, J, Aleshnick, M, Larman, B, Sukupolvi-Petty, S, Sevvana, M, Miller, A.S, Klose, T, Zheng, A, Kielian, M, Kuhn, R.J, Diamond, M.S, Pierson, T.C. | | Deposit date: | 2022-12-05 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (10.2 Å) | | Cite: | prM-reactive antibodies reveal a role for partially mature virions in dengue virus pathogenesis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7Q8Z
 
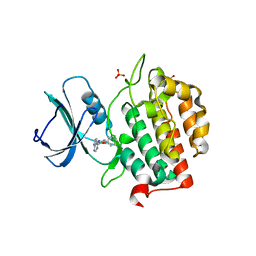 | | Crystal structure of TTBK2 in complex with VNG1.33 (compound 27) | | Descriptor: | PHOSPHATE ION, Tau-tubulin kinase 2, ~{N}-(4-phenoxyphenyl)-7~{H}-pyrrolo[2,3-d]pyrimidin-4-amine | | Authors: | Chaikuad, A, Nozal, V, Martinez, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-11 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | TDP-43 Modulation by Tau-Tubulin Kinase 1 Inhibitors: A New Avenue for Future Amyotrophic Lateral Sclerosis Therapy.
J.Med.Chem., 65, 2022
|
|
7QWS
 
 | | Structure of ribosome translating beta-tubulin in complex with TTC5 and NAC | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Jomaa, A, Gamerdinger, M, Hsieh, H, Wallisch, A, Chandrasekaran, V, Ulusoy, Z, Scaiola, A, Hegde, R, Shan, S, Ban, N, Deuerling, E. | | Deposit date: | 2022-01-25 | | Release date: | 2022-03-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting.
Science, 375, 2022
|
|
7Q90
 
 | | Crystal structure of TTBK2 in complex with VNG1.63 (compound 32) | | Descriptor: | PHOSPHATE ION, Tau-tubulin kinase 2, ~{N}-[4-(4-methoxyphenoxy)phenyl]-7~{H}-pyrrolo[2,3-d]pyrimidin-4-amine | | Authors: | Chaikuad, A, Nozal, V, Martinez, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-11 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | TDP-43 Modulation by Tau-Tubulin Kinase 1 Inhibitors: A New Avenue for Future Amyotrophic Lateral Sclerosis Therapy.
J.Med.Chem., 65, 2022
|
|
6MYO
 
 | | Avian mitochondrial complex II with flutolanyl bound | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Berry, E.A, Huang, L.-S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
8CTL
 
 | | IscB and wRNA bound to Target DNA (locked state) | | Descriptor: | DNA non-target strand, DNA target strand, IscB, ... | | Authors: | Schuler, G.A, Hu, C, Ke, A. | | Deposit date: | 2022-05-16 | | Release date: | 2022-06-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for RNA-guided DNA cleavage by IscB-omega RNA and mechanistic comparison with Cas9.
Science, 376, 2022
|
|
6MYU
 
 | | Avian mitochondrial complex II crystallized in the presence of HQNO | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Berry, E.A, Huang, L.-S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
6PIU
 
 | |
6PIY
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-2 (NR02-61) | | Descriptor: | 1,2-ETHANEDIOL, 1-methylcyclobutyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
7QZD
 
 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikF with an engineered HMA domain of Pikp-1 (Pikp-SNK-EKE) from rice (Oryza sativa) | | Descriptor: | Avr-Pik, Resistance protein Pikp-1 | | Authors: | Maidment, J.H.R, Franceschetti, M, Longya, A, Banfield, M.J. | | Deposit date: | 2022-01-31 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Effector target-guided engineering of an integrated domain expands the disease resistance profile of a rice NLR immune receptor.
Elife, 12, 2023
|
|
6PCI
 
 | | EBOV GPdMuc (Makona) in complex with rEBOV-520 and rEBOV-548 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Virion spike glycoprotein, Virion spike glycoprotein,Virion spike glycoprotein,Ebola Virus (Makona) GP2, ... | | Authors: | Ward, A.B, Murin, C.D, Alkutkar, T. | | Deposit date: | 2019-06-17 | | Release date: | 2020-03-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Analysis of a Therapeutic Antibody Cocktail Reveals Determinants for Cooperative and Broad Ebolavirus Neutralization.
Immunity, 52, 2020
|
|
6WLQ
 
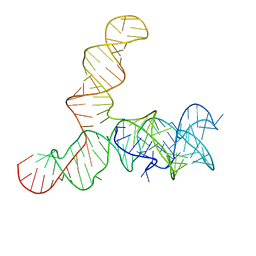 | | Apo SAM-IV riboswitch models, 4.7 Angstrom resolution | | Descriptor: | RNA (119-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6PD3
 
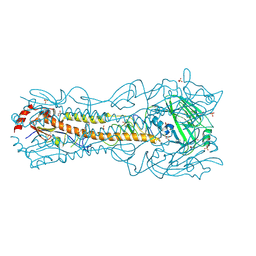 | | Crystal Structure of a H5N1 influenza virus hemagglutinin at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hemagglutinin, ... | | Authors: | Antanasijevic, A, Durst, M.A, Lavie, A, Caffrey, M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of a pH sensor in Influenza hemagglutinin using X-ray crystallography.
J.Struct.Biol., 209, 2020
|
|
6NB4
 
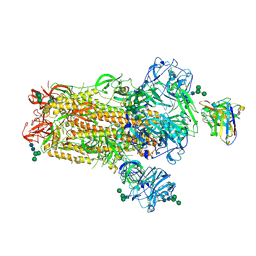 | | MERS-CoV S complex with human neutralizing LCA60 antibody Fab fragment (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LCA60 heavy chain, ... | | Authors: | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6PDJ
 
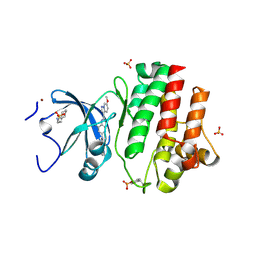 | | Tyrosine-protein kinase LCK bound to Compound 11 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, N-{4-[(6-methoxypyrazolo[1,5-a]pyridine-3-carbonyl)amino]-3-methylphenyl}-1-methyl-1H-indazole-3-carboxamide, NICKEL (II) ION, ... | | Authors: | Critton, D.A. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of Pyridazinone and Pyrazolo[1,5-a]pyridine Inhibitors of C-Terminal Src Kinase.
Acs Med.Chem.Lett., 10, 2019
|
|
6NDL
 
 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with a sulfonamide inhibitor | | Descriptor: | 1-[4-(6-aminopurin-9-yl)butylsulfamoyl]-3-[4-[(4~{S})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]butyl]urea, Biotin Protein Ligase, GLYCEROL | | Authors: | Marshall, A.C, Polyak, S.W, Bruning, J.B, Lee, K. | | Deposit date: | 2018-12-13 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sulfonamide-Based Inhibitors of Biotin Protein Ligase as New Antibiotic Leads.
Acs Chem.Biol., 14, 2019
|
|
6PEG
 
 | | MIF with a allosteric inhibitor | | Descriptor: | 4-amino-5-hydroxy-6-[(E)-(3-{[3-(2-methylpropanoyl)pyrazolo[1,5-a]pyridin-2-yl]methyl}phenyl)diazenyl]naphthalene-1,3-disulfonic acid, ACETATE ION, GLYCEROL, ... | | Authors: | Asojo, O.A. | | Deposit date: | 2019-06-20 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of Macrophage Migration Inhibitory Factor by a Chimera of Two Allosteric Binders.
Acs Med.Chem.Lett., 11, 2020
|
|
6NUL
 
 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN MUTANT: N137A REDUCED (150K) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | Deposit date: | 1997-01-09 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
5LZB
 
 | | Structure of SelB-Sec-tRNASec bound to the 70S ribosome in the initial binding state (IB) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
6PK3
 
 | |
6NFP
 
 | | 1.7 Angstrom Resolution Crystal Structure of Arginase from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | 1,2-ETHANEDIOL, Arginase, CHLORIDE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Evdokimova, E, Grimshaw, S, Kwon, K, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-20 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom Resolution Crystal Structure of Arginase from Bacillus subtilis subsp. subtilis str. 168
To Be Published
|
|
6P9D
 
 | | Crystal Structure of Pseudomonas aeruginosa D-Arginine Dehydrogenase Y249F variant with FAD - Yellow fraction | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FAD-dependent catabolic D-arginine dehydrogenase DauA, GLYCEROL | | Authors: | Reis, R.A.G, Iyer, A, Agniswamy, J, Gannavaram, S, Weber, I, Gadda, G. | | Deposit date: | 2019-06-10 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.329 Å) | | Cite: | A Single-Point Mutation in d-Arginine Dehydrogenase Unlocks a Transient Conformational State Resulting in Altered Cofactor Reactivity.
Biochemistry, 60, 2021
|
|
6NCP
 
 | | Crystal structure of HIV-1 broadly neutralizing antibody ACS202 | | Descriptor: | ACS202 Fab heavy chain, ACS202 Fab light chain, GLYCINE, ... | | Authors: | Yuan, M, Wilson, I.A. | | Deposit date: | 2018-12-11 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Conformational Plasticity in the HIV-1 Fusion Peptide Facilitates Recognition by Broadly Neutralizing Antibodies.
Cell Host Microbe, 25, 2019
|
|
