2J6I
 
 | |
7SQC
 
 | | Ciliary C1 central pair apparatus isolated from Chlamydomonas reinhardtii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CPC1, Calmodulin, ... | | Authors: | Gui, M, Wang, X, Dutcher, S.K, Brown, A, Zhang, R. | | Deposit date: | 2021-11-05 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ciliary central apparatus structure reveals mechanisms of microtubule patterning.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8IA6
 
 | |
7TFS
 
 | | Cryo-EM of the OmcE nanowires from Geobacter sulfurreducens | | Descriptor: | Cytochrome c, HEME C | | Authors: | Wang, F, Mustafa, K, Chan, C.H, Joshi, K, Hochbaum, A.I, Bond, D.R, Egelman, E.H. | | Deposit date: | 2022-01-07 | | Release date: | 2022-05-04 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of an extracellular Geobacter OmcE cytochrome filament reveals tetrahaem packing.
Nat Microbiol, 7, 2022
|
|
8IJ6
 
 | | Crystal structure of alcohol dehydrogenase from Burkholderia gladioli with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative short-chain dehydrogenases/reductase family protein | | Authors: | Han, X, Mei, Z.L, Liu, W.D, Sun, Z.T, Ma, J.A. | | Deposit date: | 2023-02-26 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of alcohol dehydrogenase from Burkholderia gladioli with NADP
To Be Published
|
|
8IJ8
 
 | |
8IJ7
 
 | |
2IDL
 
 | | Crystal Structure of Conserved Protein of Unknown Function from Streptococcus pneumoniae | | Descriptor: | GLYCEROL, Hypothetical protein, SODIUM ION | | Authors: | Nocek, B, Wu, R, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-15 | | Release date: | 2006-10-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of conserved hypothetical protein from Streptococcus pneumoniae TIGR4
To be Published
|
|
6JAF
 
 | | Crystal structure of Trypanosoma brucei gambiense glycerol kinase complex with PPi (pyrophosphatase reaction) | | Descriptor: | GLYCEROL, Glycerol kinase, PYROPHOSPHATE 2- | | Authors: | Balogun, E.O, Chishima, T, Ichinose, M, Inaoka, D.K, Kido, Y, Ibrahim, B, Bringaud, F, de Koning, H, McKerrow, J.H, Watanabe, Y, Nozaki, T, Michels, P.A.M, Harada, S, Kita, K, Shiba, T. | | Deposit date: | 2019-01-24 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Glycerol Kinase of African Human Trypanosomes Possesses a Pyrophosphatase Activity.
To Be Published
|
|
8IJG
 
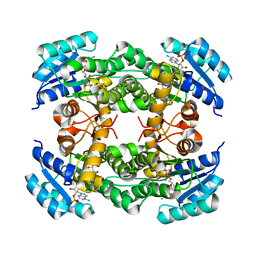 | | Crystal structure of alcohol dehydrogenase M5 from Burkholderia gladioli with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative short-chain dehydrogenases/reductase family protein | | Authors: | Han, X, Mei, Z.L, Liu, W.D, Sun, Z.T, Ma, J.A. | | Deposit date: | 2023-02-27 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of alcohol dehydrogenase from Burkholderia gladioli with NADP
To Be Published
|
|
2IDY
 
 | | NMR Structure of the SARS-CoV non-structural protein nsp3a | | Descriptor: | NSP3 | | Authors: | Serrano, P, Almeida, M.S, Johnson, M.A, Horst, R, Herrmann, T, Joseph, J, Saikatendu, K, Subramanian, V, Stevens, R.C, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-09-15 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
6IXV
 
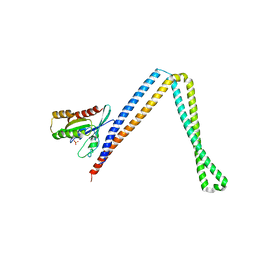 | | Crystal structure of SH3BP5-Rab11a | | Descriptor: | PHOSPHATE ION, Ras-related protein Rab-11A, SH3 domain-binding protein 5 | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | Deposit date: | 2018-12-12 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of guanine nucleotide exchange for Rab11 by SH3BP5.
Life Sci Alliance, 2, 2019
|
|
4I1Z
 
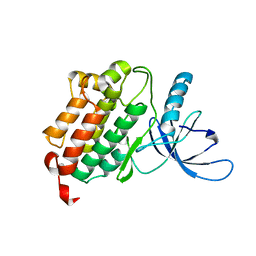 | | Crystal structure of the monomeric (V948R) form of the gefitinib/erlotinib resistant EGFR kinase domain L858R+T790M | | Descriptor: | Epidermal growth factor receptor | | Authors: | Gajiwala, K.S, Feng, J, Ferre, R, Ryan, K, Brodsky, O, Stewart, A. | | Deposit date: | 2012-11-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights into the Aberrant Activity of Mutant EGFR Kinase Domain and Drug Recognition.
Structure, 21, 2013
|
|
8TJP
 
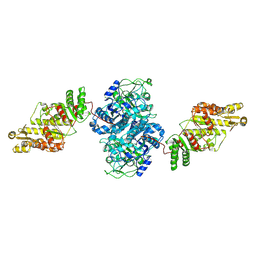 | | KS-AT core of 6-deoxyerythronolide B synthase (DEBS) Module 3 crosslinked with its elongation ACP partner | | Descriptor: | EryAII | | Authors: | Cogan, D.P, Soohoo, A.M, Chen, M, Brodsky, K.L, Liu, Y, Khosla, C. | | Deposit date: | 2023-07-23 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural Basis for Intermodular Communication in Assembly-Line Polyketide Biosynthesis
To Be Published
|
|
8K4O
 
 | |
7ZWD
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U5 snRNA promoter (CC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZX7
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (CC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZXE
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (OC) | | Descriptor: | Non-template strand, TATA-box-binding protein, Template strand, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4QXU
 
 | | Novel Inhibition Mechanism of Membrane Metalloprotease by an Exosite-Swiveling Conformational antibody | | Descriptor: | Matrix metalloproteinase-14, SULFATE ION, anti_MT1-MMP Heavy chain, ... | | Authors: | Udi, Y, Grossman, M, Solomonov, I, Dym, O, Rozenberg, H, Moreno, v, Cuiniasse, P, Dive, V, Arroyo, A.G, Sagi, I, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2014-07-22 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibition mechanism of membrane metalloprotease by an exosite-swiveling conformational antibody.
Structure, 23, 2015
|
|
7ZX8
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (OC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4RAF
 
 | | Crystal structure of PP2Ca-D38A | | Descriptor: | MANGANESE (II) ION, Protein phosphatase 1A | | Authors: | Pan, C, Tang, J.Y, Xu, Y.F, Xiao, P, Liu, H.D, Wang, H.A, Wang, W.B, Meng, F.G, Yu, X, Sun, J.P. | | Deposit date: | 2014-09-10 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | The catalytic role of the M2 metal ion in PP2C alpha
Sci Rep, 5, 2015
|
|
1T2A
 
 | | Crystal structure of human GDP-D-mannose 4,6-dehydratase | | Descriptor: | GDP-mannose 4,6 dehydratase, GUANOSINE-5'-DIPHOSPHATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Walker, J.R, Vedadi, M, Sharma, S, Houston, S, Wasney, G, Loppnau, P, Sundstrom, M, Arrowsmith, C, Edwards, A, Oppermann, U. | | Deposit date: | 2004-04-20 | | Release date: | 2004-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure and Biophysical Characterization of Human GDP-D-mannose 4,6-dehydratase
To be Published
|
|
3E5Z
 
 | | X-Ray structure of the putative gluconolactonase in protein family PF08450. Northeast Structural Genomics Consortium target DrR130. | | Descriptor: | MAGNESIUM ION, putative Gluconolactonase | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Wang, D, Mao, L, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Tong, S.N, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-08-14 | | Release date: | 2008-09-30 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | X-Ray structure of the putative gluconolactonase in protein family PF08450. Northeast Structural Genomics Consortium target DrR130.
To be Published
|
|
3E7H
 
 | |
3EAG
 
 | |
