6U5B
 
 | | CryoEM Structure of Pyocin R2 - precontracted - baseplate | | Descriptor: | Glue PA0627, Ripcord PA0626, Sheath Initiator PA0617, ... | | Authors: | Ge, P, Avaylon, J, Scholl, D, Shneider, M.M, Browning, C, Buth, S.A, Plattner, M, Ding, K, Leiman, P.G, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2019-08-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Action of a minimal contractile bactericidal nanomachine.
Nature, 580, 2020
|
|
8C0F
 
 | | Tubulin-PTC596 complex | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-fluoranyl-2-(6-fluoranyl-2-methyl-benzimidazol-1-yl)-~{N}4-[4-(trifluoromethyl)phenyl]pyrimidine-4,6-diamine, ... | | Authors: | Prota, A.E, Muehlethaler, T, Weetall, M, Steinmetz, M.O. | | Deposit date: | 2022-12-16 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1005 Å) | | Cite: | Preclinical and Early Clinical Development of PTC596, a Novel Small-Molecule Tubulin-Binding Agent
Mol Cancer Ther, 20, 2021
|
|
5VBT
 
 | | Crystal structure of a highly specific and potent USP7 ubiquitin variant inhibitor | | Descriptor: | UBH04 | | Authors: | DONG, A, DONG, X, LIU, L, GUO, Y, LI, Y, ZHANG, W, WALKER, J.R, SIDHU, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, TONG, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-30 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of a highly specific and potent USP7 ubiquitin variant inhibitor
to be published
|
|
7K2T
 
 | |
6BYI
 
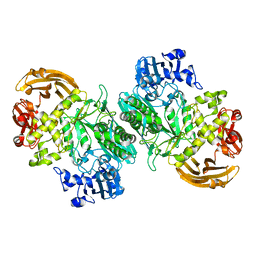 | | Crystal structure of the acid-base mutant (E477A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri | | Descriptor: | Beta-mannosidase, beta-D-mannopyranose | | Authors: | Domingues, M.N, Vieira, P.S, Morais, M.A.B, Murakami, M.T. | | Deposit date: | 2017-12-20 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of exo-beta-mannanase activity in the GH2 family.
J. Biol. Chem., 293, 2018
|
|
7MWL
 
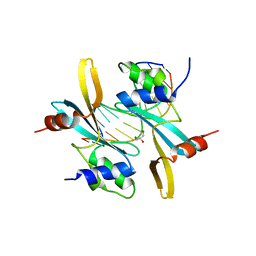 | | The TAM domain of BAZ2A in complex with a 12mer mCG DNA | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), GLYCEROL | | Authors: | Liu, K, Dong, A, Li, Y, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-17 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The TAM domain of BAZ2A in complex with a 12mer mCG DNA
To Be Published
|
|
7V0I
 
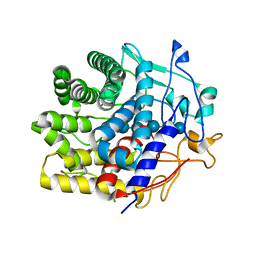 | | Crystal structure of a CelR catalytic domain active site mutant with bound cellohexaose substrate | | Descriptor: | CALCIUM ION, Glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Bingman, C.A, Kuch, N, Kutsche, M.E, Parker, A, Smith, R.W, Fox, B.G. | | Deposit date: | 2022-05-10 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Contribution of calcium ligands in substrate binding and product release in the Acetovibrio thermocellus glycoside hydrolase family 9 cellulase CelR.
J.Biol.Chem., 299, 2023
|
|
7W7Z
 
 | |
8V8K
 
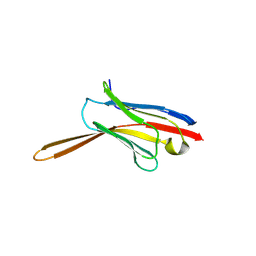 | | Crystal Structure of Nanobody NbE | | Descriptor: | Nanobody NbE | | Authors: | Koehl, A, Manglik, A, Yu, J, Kumar, A, Zhang, X, Martin, C, Raia, P, Steyaert, J, Ballet, S, Boland, A, Stoeber, M. | | Deposit date: | 2023-12-05 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis of mu-Opioid Receptor-Targeting by a Nanobody Antagonist
To Be Published
|
|
5G17
 
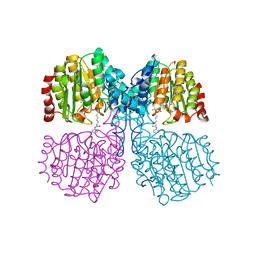 | | Bordetella Alcaligenes HDAH (T101A) bound to 9,9,9-trifluoro-8,8- dihydroxy-N-phenylnonanamide. | | Descriptor: | 9,9,9-tris(fluoranyl)-8,8-bis(oxidanyl)-~{N}-phenyl-nonanamide, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, POTASSIUM ION, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-03-23 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The thermodynamic signature of ligand binding to histone deacetylase-like amidohydrolases is most sensitive to the flexibility in the L2-loop lining the active site pocket.
Biochim. Biophys. Acta, 1861, 2017
|
|
7JRL
 
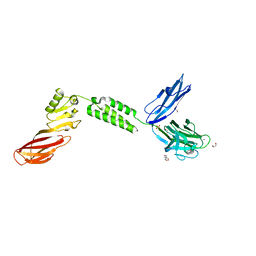 | | The structure of CBM51-2 in complex with GlcNAc and INT domains from Clostridium perfringens ZmpB | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2020-08-12 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Architecturally complex O -glycopeptidases are customized for mucin recognition and hydrolysis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1Q8X
 
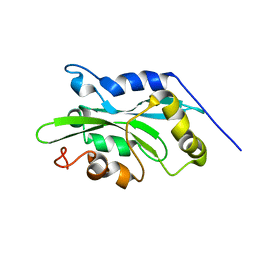 | | NMR structure of human cofilin | | Descriptor: | Cofilin, non-muscle isoform | | Authors: | Pope, B.J, Zierler-Gould, K.M, Kuhne, R, Weeds, A.G, Ball, L.J. | | Deposit date: | 2003-08-22 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human cofilin: rationalizing actin binding and pH sensitivity
J.Biol.Chem., 279, 2004
|
|
5QJD
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z240297434 | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-N-(1-methyl-1H-pyrazol-3-yl)-1,2-oxazole-5-carboxamide, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
7BIP
 
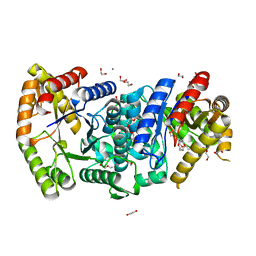 | | Crystal structure of monooxygenase RslO1 from Streptomyces bottropensis | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Zhang, L, Zuo, C, Bechthold, A, Einsle, O. | | Deposit date: | 2021-01-12 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biosynthesis of the Tricyclic Aromatic Type II Polyketide Rishirilide: New Potential Third Ring Oxygenation after Three Cyclization Steps.
Mol Biotechnol., 63, 2021
|
|
8UX9
 
 | | Asymmetric unit of the PARIS Immune Complex at 3.2 Angstrom Resolution | | Descriptor: | AriA, AriB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Burman, N.B, Henriques, W, Wilkinson, R, Graham, A, Wiedenheft, B. | | Deposit date: | 2023-11-09 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Activation of the PARIS immune complex results in tRNA cleavage and can be subverted by viral tRNAs
To Be Published
|
|
6N12
 
 | | Structure of GTPase Domain of Human Septin 7 at High Resolution | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Septin-7 | | Authors: | Brognara, G, Pereira, H.M, Brandao-Neto, J, Araujo, A.P.U, Garratt, R.C. | | Deposit date: | 2018-11-08 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Revisiting SEPT7 and the slippage of beta-strands in the septin family.
J.Struct.Biol., 207, 2019
|
|
2XNQ
 
 | |
3PFH
 
 | | X-Ray crystal structure the N,N-dimethyltransferase TylM1 from Streptomyces fradiae in complex with SAH and dTDP-Quip3N | | Descriptor: | 1,2-ETHANEDIOL, N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Carney, A.E, Holden, H.M. | | Deposit date: | 2010-10-28 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Molecular Architecture of TylM1 from Streptomyces fradiae: An N,N-Dimethyltransferase Involved in the Production of dTDP-d-mycaminose .
Biochemistry, 50, 2011
|
|
6FA0
 
 | |
7BBH
 
 | | Structure of Coronavirus Spike from Smuggled Guangdong Pangolin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Surface glycoprotein | | Authors: | Wrobel, A.G, Benton, D.J, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-12-17 | | Release date: | 2020-12-30 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and binding properties of Pangolin-CoV spike glycoprotein inform the evolution of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
5QPU
 
 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000733a | | Descriptor: | ACETATE ION, Farnesyl diphosphate synthase, N-[(4-phenyloxan-4-yl)methyl]acetamide, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
7B3J
 
 | | Dynamic complex between all-D-enantiomeric peptide D3 with wild-type amyloid precursor protein 672-726 fragment (amyloid beta 1-55) | | Descriptor: | D3 all D-enantimeric peptide, Isoform L-APP677 of Amyloid-beta precursor protein | | Authors: | Bocharov, E.V, Volynsky, P.E, Okhrimenko, I.S, Urban, A.S. | | Deposit date: | 2020-12-01 | | Release date: | 2021-01-13 | | Last modified: | 2021-12-08 | | Method: | SOLUTION NMR | | Cite: | All - d - Enantiomeric Peptide D3 Designed for Alzheimer's Disease Treatment Dynamically Interacts with Membrane-Bound Amyloid-beta Precursors.
J.Med.Chem., 64, 2021
|
|
5FYD
 
 | | Structural and biochemical insights into 7beta-hydroxysteroid dehydrogenase stereoselectivity | | Descriptor: | GLYCEROL, OXIDOREDUCTASE, SHORT CHAIN DEHYDROGENASE/REDUCTASE FAMILY PROTEIN | | Authors: | Savino, S, Ferrandi, E, Forneris, F, Rovida, S, Riva, S, Monti, D, Mattevi, A. | | Deposit date: | 2016-03-07 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Insights Into 7Beta-Hydroxysteroid Dehydrogenase Stereoselectivity.
Proteins, 84, 2016
|
|
5X00
 
 | |
4Z45
 
 | | Structure of OBP3 from the currant-lettuce aphid Nasonovia ribisnigri | | Descriptor: | Odorant-binding protein NribOBP3 | | Authors: | Northey, T, Venthur, H, De Biasio, F, Chauviac, F.-X, Cole, A.R, Field, L.M, Zhou, J.-J, Keep, N.H. | | Deposit date: | 2015-04-01 | | Release date: | 2016-04-13 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal Structures and Binding Dynamics of Odorant-Binding Protein 3 from two aphid species Megoura viciae and Nasonovia ribisnigri.
Sci Rep, 6, 2016
|
|
