4E40
 
 | | The haptoglobin-hemoglobin receptor of Trypanosoma congolense | | Descriptor: | Putative uncharacterized protein | | Authors: | Higgins, M.K, Tkachenko, O, Brown, A, Reed, J, Carrington, M. | | Deposit date: | 2012-03-11 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the trypanosome haptoglobin-hemoglobin receptor and implications for nutrient uptake and innate immunity.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6XWU
 
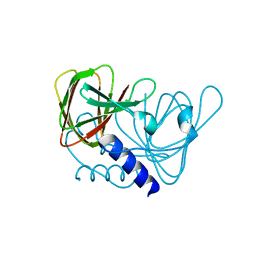 | | Crystal structure of drosophila melanogaster CENP-C cumin domain | | Descriptor: | RE68959p | | Authors: | Jeyaprakash, A.A, Medina-Pritchard, B, Lazou, V, Zou, J, Byron, O, Abad, M.A, Rappsilber, J, Heun, P. | | Deposit date: | 2020-01-24 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural basis for centromere maintenance by Drosophila CENP-A chaperone CAL1.
Embo J., 39, 2020
|
|
2Y6S
 
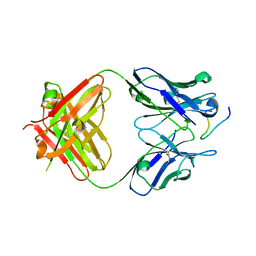 | | Structure of an Ebolavirus-protective antibody in complex with its mucin-domain linear epitope | | Descriptor: | ENVELOPE GLYCOPROTEIN, HEAVY CHAIN, LIGHT CHAIN | | Authors: | Olal, D.O, Kuehne, A, Lee, J.E, Bale, S, Dye, J.M, Saphire, E.O. | | Deposit date: | 2011-01-25 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an Ebola Virus-Protective Antibody in Complex with its Mucin-Domain Linear Epitope.
J.Virol., 86, 2012
|
|
7R8M
 
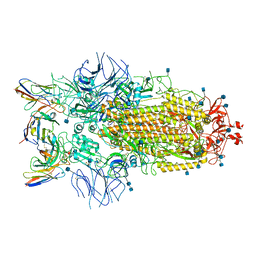 | | Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C032 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | DeLaitsch, A.T, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-06-26 | | Release date: | 2021-08-04 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
4YVA
 
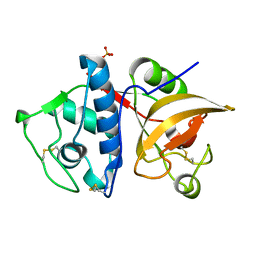 | | Cathepsin K co-crystallized with actinomycetes extract | | Descriptor: | Cathepsin K, SULFATE ION | | Authors: | Aguda, A.H, Nguyen, N.T, Bromme, D, Brayer, G.D. | | Deposit date: | 2015-03-19 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Affinity Crystallography: A New Approach to Extracting High-Affinity Enzyme Inhibitors from Natural Extracts.
J.Nat.Prod., 79, 2016
|
|
6BKF
 
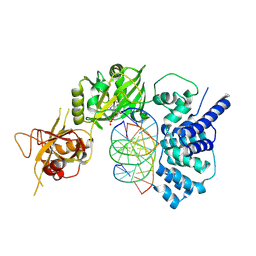 | | Lysyl-adenylate form of human LigIV catalytic domain with bound DNA substrate in open conformation | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*C)-3'), DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*GP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Moon, A.F, Tumbale, P.P, Schellenberg, M.J, Williams, R.S, Williams, J.G, Kunkel, T.A, Pedersen, L.C, Bebenek, B. | | Deposit date: | 2017-11-08 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structures of DNA-bound human ligase IV catalytic core reveal insights into substrate binding and catalysis.
Nat Commun, 9, 2018
|
|
5CNS
 
 | | Crystal structure of the dATP inhibited E. coli class Ia ribonucleotide reductase complex bound to CDP and dATP at 2.97 Angstroms resolution | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, CYTIDINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, P.Y.-T, Zimanyi, C.M, Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-07-18 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.975 Å) | | Cite: | Molecular basis for allosteric specificity regulation in class Ia ribonucleotide reductase from Escherichia coli.
Elife, 5, 2016
|
|
5G4F
 
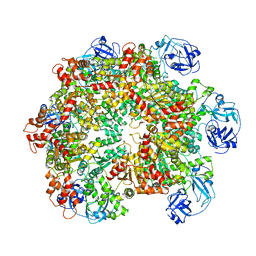 | | Structure of the ADP-bound VAT complex | | Descriptor: | VCP-LIKE ATPASE | | Authors: | Huang, R, Ripstein, Z.A, Augustyniak, R, Lazniewski, M, Ginalski, K, Kay, L.E, Rubinstein, J.L. | | Deposit date: | 2016-05-12 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Unfolding the Mechanism of the Aaa+ Unfoldase Vat by a Combined Cryo-Em, Solution NMR Study.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3ZRC
 
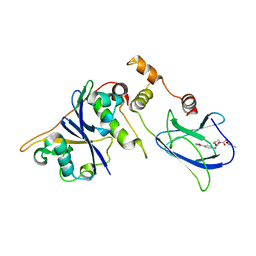 | | pVHL54-213-EloB-EloC complex (4R)-4-HYDROXY-1-[(3-METHYLISOXAZOL-5-YL)ACETYL]-N-[4-(1,3-OXAZOL-5-YL)BENZYL]-L-PROLINAMIDE bound | | Descriptor: | (4R)-4-HYDROXY-1-[(3-METHYLISOXAZOL-5-YL)ACETYL]-N-[4-(1,3-OXAZOL-5-YL)BENZYL]-L-PROLINAMIDE, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 1, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 2, ... | | Authors: | Van Molle, I, Buckley, D.L, Crews, C.M, Ciulli, A. | | Deposit date: | 2011-06-15 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Targeting the Von Hippel-Lindau E3 Ubiquitin Ligase Using Small Molecules to Disrupt the Vhl/Hif-1Alpha Interaction
J.Am.Chem.Soc., 134, 2012
|
|
4NO6
 
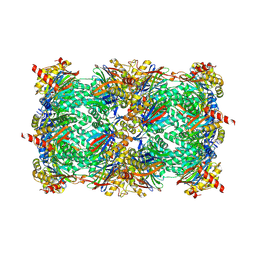 | | yCP in complex with Z-Leu-Leu-Leu-vinylsulfone | | Descriptor: | MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(3S)-5-methyl-1-(methylsulfonyl)hexan-3-yl]-L-leucinamide, Probable proteasome subunit alpha type-7, ... | | Authors: | Stein, M.L, Cui, H, Beck, P, Dubiella, C, Voss, C, Krueger, A, Schmidt, B, Groll, M. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Systematic Comparison of Peptidic Proteasome Inhibitors Highlights the alpha-Ketoamide Electrophile as an Auspicious Reversible Lead Motif.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5JXF
 
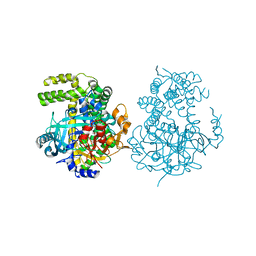 | | Crystal structure of Flavobacterium psychrophilum DPP11 in complex with dipeptide Arg-Asp | | Descriptor: | ARGININE, ASPARTIC ACID, Asp/Glu-specific dipeptidyl-peptidase, ... | | Authors: | Bezerra, G.A, Fedosyuk, S, Ohara-Nemoto, Y, Nemoto, T.K, Djinovic-Carugo, K. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
4NO1
 
 | | yCP in complex with Z-Leu-Leu-Leu-B(OH)2 | | Descriptor: | MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(1R)-1-(dihydroxyboranyl)-3-methylbutyl]-L-leucinamide, Probable proteasome subunit alpha type-7, ... | | Authors: | Stein, M.L, Cui, H, Beck, P, Dubiella, C, Voss, C, Krueger, A, Schmidt, B, Groll, M. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systematic Comparison of Peptidic Proteasome Inhibitors Highlights the alpha-Ketoamide Electrophile as an Auspicious Reversible Lead Motif.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1ON0
 
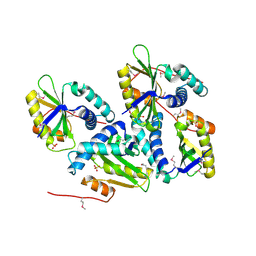 | | Crystal Structure of Putative Acetyltransferase (YycN) from Bacillus subtilis, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET SR144 | | Descriptor: | CHLORIDE ION, SULFATE ION, YycN protein | | Authors: | Forouhar, F, Shen, J, Kuzin, A, Chiang, Y, Xiao, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-26 | | Release date: | 2003-03-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Putative Acetyltransferase (YycN) from Bacillus subtilis
To be Published
|
|
5TPA
 
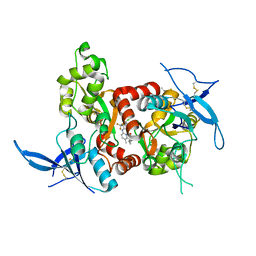 | | Structure of the human GluN1/GluN2A LBD in complex with compound 9 (GNE3500) | | Descriptor: | (1R,2R)-2-(2-{[5-chloro-3-(trifluoromethyl)-1H-pyrazol-1-yl]methyl}-7-methyl-4-oxo-4H-pyrido[1,2-a]pyrimidin-6-yl)cyclopropane-1-carbonitrile, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | GluN2A-Selective Pyridopyrimidinone Series of NMDAR Positive Allosteric Modulators with an Improved in Vivo Profile.
ACS Med Chem Lett, 8, 2017
|
|
6F55
 
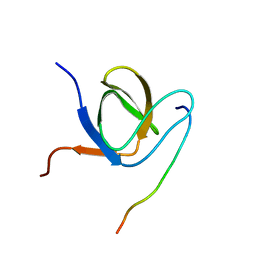 | | Complex structure of PACSIN SH3 domain and TRPV4 proline rich region | | Descriptor: | PACSIN 3, PRR | | Authors: | Glogowski, N.A, Goretzki, B, Diehl, E, Duchardt-Ferner, E, Hacker, C, Hellmich, U.A. | | Deposit date: | 2017-11-30 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of TRPV4 N Terminus Interaction with Syndapin/PACSIN1-3 and PIP2.
Structure, 26, 2018
|
|
4YV7
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM MYCOBACTERIUM SMEGMATIS (MSMEI_3018, TARGET EFI-511327) WITH BOUND GLYCEROL | | Descriptor: | GLYCEROL, Periplasmic binding protein/LacI transcriptional regulator | | Authors: | Vetting, M.W, Patskovsky, Y, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-19 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM MYCOBACTERIUM SMEGMATIS (MSMEI_3018, TARGET EFI-511327) WITH BOUND GLYCEROL
To be published
|
|
7TMW
 
 | | Cryo-EM structure of the relaxin receptor RXFP1 in complex with heterotrimeric Gs | | Descriptor: | Camelid antibody VHH fragment Nb35, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Erlandson, S.C, Rawson, S, Kruse, A.C. | | Deposit date: | 2022-01-20 | | Release date: | 2023-02-15 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The relaxin receptor RXFP1 signals through a mechanism of autoinhibition.
Nat.Chem.Biol., 19, 2023
|
|
3RWC
 
 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*17-IW9 | | Descriptor: | Beta-2-microglobulin, Major histocompatibility complex class I, Nef IW9 peptide from Protein Nef | | Authors: | Wu, Y, Gao, F, Liu, J, Qi, J.X, Price, D.A, Gao, G.F. | | Deposit date: | 2011-05-09 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structural basis of diverse peptide accommodation by the rhesus macaque MHC class I molecule Mamu-B*17: insights into immune protection from simian immunodeficiency virus
J.Immunol., 187, 2011
|
|
6S9L
 
 | | Designed Armadillo Repeat protein Lock1 bound to (KR)4KLSF target | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, KR4KLSF Lock1, LYS-ARG-LYS-ARG-LYS-ARG-LYS-ARG-LYS-LEU-SER-PHE | | Authors: | Ernst, P, Zosel, F, Reichen, C, Schuler, B, Pluckthun, A. | | Deposit date: | 2019-07-15 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Design of a Peptide Lock for Modular Peptide Binders.
Acs Chem.Biol., 15, 2020
|
|
6BYN
 
 | | Crystal structure of WDR5-Mb(S4) monobody complex | | Descriptor: | WD repeat-containing protein 5, WDR5-binding Monobody, Mb(S4) | | Authors: | Gupta, A, Koide, S. | | Deposit date: | 2017-12-21 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Facile target validation in an animal model with intracellularly expressed monobodies.
Nat. Chem. Biol., 14, 2018
|
|
5CG2
 
 | | Crystal structure of E. coli FabI bound to the thiocarbamoylated benzodiazaborine inhibitor 35b. | | Descriptor: | 1-hydroxy-2,3,1-benzodiazaborinine-2(1H)-carbothioamide, Enoyl-[acyl-carrier-protein] reductase [NADH] FabI, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jordan, C.A, Vey, J.L. | | Deposit date: | 2015-07-09 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystallographic insights into the structure-activity relationships of diazaborine enoyl-ACP reductase inhibitors.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5JSL
 
 | | The L16F mutant of cytochrome c prime from Alcaligenes xylosoxidans: Ferrous form | | Descriptor: | ASCORBIC ACID, Cytochrome c', HEME C, ... | | Authors: | Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2016-05-08 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Engineering proximal vs. distal heme-NO coordination via dinitrosyl dynamics: implications for NO sensor design.
Chem Sci, 8, 2017
|
|
1OQ1
 
 | |
4YUQ
 
 | | CDPK1 from Eimeria tenella in complex with inhibitor UW1354 | | Descriptor: | 1-cyclopentyl-3-(1H-pyrrolo[2,3-b]pyridin-5-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, CALCIUM ION, Calmodulin-like domain protein kinase | | Authors: | Merritt, E.A. | | Deposit date: | 2015-03-19 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CDPK is a druggable target in the apicomplexan parasite Eimeria
to be published
|
|
6P1W
 
 | | Pre-catalytic ternary complex of human DNA Polymerase Mu with 1-nt gapped substrate containing undamaged template dG and bound incoming CMPCPP | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, ... | | Authors: | Kaminski, A.M, Pedersen, L.C, Bebenek, K, Chiruvella, K.K, Ramsden, D.A, Kunkel, T.A. | | Deposit date: | 2019-05-20 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Unexpected behavior of DNA polymerase Mu opposite template 8-oxo-7,8-dihydro-2'-guanosine.
Nucleic Acids Res., 47, 2019
|
|
