6TDZ
 
 | | Cryo-EM structure of Euglena gracilis mitochondrial ATP synthase, OSCP/F1/c-ring, rotational state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, FRAGMENT OF TRITON X-100, ... | | Authors: | Muhleip, A, Amunts, A. | | Deposit date: | 2019-11-10 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structure of a mitochondrial ATP synthase with bound native cardiolipin.
Elife, 8, 2019
|
|
8C5I
 
 | | Cyanide dihydratase from Bacillus pumilus C1 variant - Q86R,H305K,H308K,H323K | | Descriptor: | Cyanide dihydratase | | Authors: | Mulelu, A.E, Reitz, J, van Rooyen, J, Scheffer, M, Frangakis, A.S, Dlamini, L.S, Woodward, J.D, Benedik, M.J, Sewell, B.T. | | Deposit date: | 2023-01-09 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | The Role of Histidine Residues in the Oligomerization of Cyanide Dihydratase from Bacillus pumilus C1
To Be Published
|
|
7JSS
 
 | |
8SBM
 
 | | Crystal structure of the wild-type Catalytic ATP-binding domain of Mtb DosS | | Descriptor: | 1,2-ETHANEDIOL, GAF domain-containing protein, SODIUM ION, ... | | Authors: | Larson, G, Shi, K, Aihara, H, Bhagi-Damodaran, A. | | Deposit date: | 2023-04-03 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Understanding ATP Binding to DosS Catalytic Domain with a Short ATP-Lid.
Biochemistry, 62, 2023
|
|
3MBY
 
 | | Ternary complex of DNA Polymerase BETA with template base A and 8oxodGTP in the active site with a dideoxy terminated primer | | Descriptor: | 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Hou, E.W, Pedersen, L.C, Prasad, R, Wilson, S.H. | | Deposit date: | 2010-03-26 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutagenic conformation of 8-oxo-7,8-dihydro-2'-dGTP in the confines of a DNA polymerase active site.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7JXC
 
 | | Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology | | Descriptor: | NONAETHYLENE GLYCOL, S2H14 antigen-binding (Fab) fragment | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
5G09
 
 | | The crystal structure of a S-selective transaminase from Bacillus megaterium bound with R-alpha-methylbenzylamine | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, TETRAETHYLENE GLYCOL, ... | | Authors: | van Oosterwijk, N, Willies, S, Hekelaar, J, Terwisscha van Scheltinga, A.C, Turner, N.J, Dijkstra, B.W. | | Deposit date: | 2016-03-17 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Substrate Range and Enantioselectivity of Two S-Selective Omega- Transaminases
Biochemistry, 55, 2016
|
|
5Y85
 
 | |
6C83
 
 | | Structure of Aurora A (122-403) bound to inhibitory Monobody Mb2 and AMPPCP | | Descriptor: | Aurora kinase A, Mb2 Monobody, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Hoemberger, M, Kutter, S, Zorba, A, Nguyen, V, Shohei, A, Shohei, K, Kern, D. | | Deposit date: | 2018-01-24 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Allosteric modulation of a human protein kinase with monobodies.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4YD7
 
 | | Endothiapepsin in complex with fragment 255 | | Descriptor: | 2-(imidazo[1,2-a]pyridin-2-yl)-N-phenylacetamide, ACETATE ION, Endothiapepsin, ... | | Authors: | Stieler, M, Heine, A, Klebe, G. | | Deposit date: | 2015-02-21 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
7MWV
 
 | |
6MI6
 
 | | STRUCTURE OF CHEA DOMAIN P4 IN COMPLEX WITH AN ADP ANALOG | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy{[(1-hydroxy-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl]disulfanyl}phosphoryl]oxy}phosphoryl]adenosine, ADENOSINE-5'-DIPHOSPHATE, Chemotaxis protein CheA, ... | | Authors: | Crane, B.R, Muok, A.R, Chua, T.K, Le, H. | | Deposit date: | 2018-09-19 | | Release date: | 2018-12-05 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Nucleotide Spin Labeling for ESR Spectroscopy of ATP-Binding Proteins.
Appl.Magn.Reson., 49, 2018
|
|
3M17
 
 | | Crystal structure of human FcRn with a monomeric peptide inhibitor | | Descriptor: | Beta-2-microglobulin, IgG receptor FcRn large subunit p51, monomeric peptide inhibitor | | Authors: | Mezo, A.R, Sridhar, V, Badger, J, Sakorafas, P, Nienaber, V. | | Deposit date: | 2010-03-04 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray crystal structures of monomeric and dimeric peptide inhibitors in complex with the human neonatal Fc receptor, FcRn.
J.Biol.Chem., 285, 2010
|
|
6XSX
 
 | |
8R8C
 
 | |
7MWU
 
 | |
6XI7
 
 | | Crystal Structure of wild-type KRAS (GMPPNP-bound) in complex with RAS-binding domain (RBD) and cysteine-rich domain (CRD) of RAF1/CRAF (crystal form I) | | Descriptor: | CHLORIDE ION, Isoform 2B of GTPase KRas, MAGNESIUM ION, ... | | Authors: | Chan, A.H, Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2020-06-19 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation.
Nat Commun, 12, 2021
|
|
4YUR
 
 | | Crystal Structure of Plk4 Kinase Domain Bound to Centrinone | | Descriptor: | 2-({2-fluoro-4-[(2-fluoro-3-nitrobenzyl)sulfonyl]phenyl}sulfanyl)-5-methoxy-N-(3-methyl-1H-pyrazol-5-yl)-6-(morpholin-4-yl)pyrimidin-4-amine, Serine/threonine-protein kinase PLK4 | | Authors: | Shiau, A.K, Motamedi, A. | | Deposit date: | 2015-03-19 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4.
Science, 348, 2015
|
|
6MKU
 
 | | Crystal structure of the periplasmic Lysine-, Arginine-, Ornithine-binding protein (LAO) D11A mutant from Salmonella typhimurium complexed with arginine | | Descriptor: | ACETATE ION, ARGININE, GLYCEROL, ... | | Authors: | Romero-Romero, S, Vergara, R, Espinoza-Perez, G, Rodriguez-Romero, A. | | Deposit date: | 2018-09-26 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.727 Å) | | Cite: | The interplay of protein-ligand and water-mediated interactions shape affinity and selectivity in the LAO binding protein.
Febs J., 287, 2020
|
|
6TIG
 
 | | Structure of the N terminal domain of Bc2L-C lectin (1-131) in complex with Globo H (H-type 3) antigen | | Descriptor: | Lectin, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose | | Authors: | Varrot, A, Bermeo, R. | | Deposit date: | 2019-11-22 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | BC2L-C N-Terminal Lectin Domain Complexed with Histo Blood Group Oligosaccharides Provides New Structural Information.
Molecules, 25, 2020
|
|
5KGD
 
 | |
7TBE
 
 | | Crystal structure of Plasmepsin X from Plasmodium vivax in complex with WM4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(R)-[(2E,4S)-2-imino-4-methyl-6-oxo-4-(propan-2-yl)-1,3-diazinan-1-yl](phenyl)methyl]-N-[(1S)-1-phenylethyl]benzamide, Plasmepsin X, ... | | Authors: | Hodder, A.N, Christensen, J.B, Scally, S.W, Cowman, A.F. | | Deposit date: | 2021-12-21 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Basis for drug selectivity of plasmepsin IX and X inhibition in Plasmodium falciparum and vivax.
Structure, 30, 2022
|
|
8BYK
 
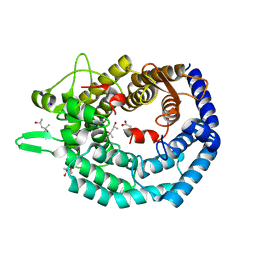 | | The structure of MadC from Clostridium maddingley reveals new insights into class I lanthipeptide cyclases | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Knospe, C.V, Kamel, M, Spitz, O, Hoeppner, A, Galle, S, Reiners, J, Kedrov, A, Smits, S.H, Schmitt, L. | | Deposit date: | 2022-12-13 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of MadC from Clostridium maddingley reveals new insights into class I lanthipeptide cyclases.
Front Microbiol, 13, 2022
|
|
6F7C
 
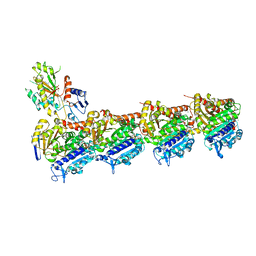 | | TUBULIN-Compound 12 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,4,5-trimethoxy-~{N}-[(~{E})-naphthalen-1-ylmethylideneamino]benzamide, CALCIUM ION, ... | | Authors: | Muehlethaler, T, Prota, A.E, Steinmetz, M.O. | | Deposit date: | 2017-12-08 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structural Basis of Colchicine-Site targeting Acylhydrazones active against Multidrug-Resistant Acute Lymphoblastic Leukemia.
Iscience, 21, 2019
|
|
4RK4
 
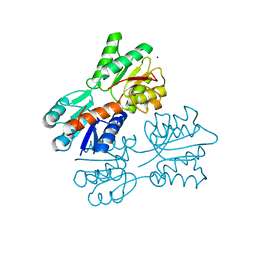 | | Crystal structure of LacI family transcriptional regulator from Lactobacillus casei, Target EFI-512911, with bound glucose | | Descriptor: | SODIUM ION, Transcriptional regulator, LacI family, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-11 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystal Structure of LacI Transcriptional Regulator Lsei_2103 from Lactobacillus casei, Target EFI-512911
To be Published
|
|
