3EFG
 
 | | Structure of SlyX protein from Xanthomonas campestris pv. campestris str. ATCC 33913 | | Descriptor: | 1,2-ETHANEDIOL, Protein slyX homolog | | Authors: | Cuff, M.E, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-08 | | Release date: | 2008-12-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of SlyX protein from Xanthomonas campestris pv. campestris str. ATCC 33913
TO BE PUBLISHED
|
|
3EH5
 
 | | Structure of the reduced form of cytochrome ba3 oxidase from Thermus thermophilus | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, Cytochrome c oxidase subunit 1, ... | | Authors: | Liu, B, Chen, Y, Doukov, T, Soltis, S.M, Stout, D, Fee, J.A. | | Deposit date: | 2008-09-11 | | Release date: | 2009-04-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Combined microspectrophotometric and crystallographic examination of chemically reduced and X-ray radiation-reduced forms of cytochrome ba3 oxidase from Thermus thermophilus: structure of the reduced form of the enzyme.
Biochemistry, 48, 2009
|
|
6IC6
 
 | | Human cathepsin-C in complex with cyclopropyl peptidyl nitrile inhibitor 1 | | Descriptor: | (2~{S})-~{N}-[(1~{R},2~{R})-1-(aminomethyl)-2-[4-[4-(trifluoromethyl)phenyl]phenyl]cyclopropyl]-2-azanyl-butanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hakansson, M, Logan, D.T, Korkmaz, B, Lesner, A, Wysocka, M, Gieldon, A, Gauthier, F, Jenne, D, Lauritzen, C, Pedersen, J. | | Deposit date: | 2018-12-02 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Structure-based design and in vivo anti-arthritic activity evaluation of a potent dipeptidyl cyclopropyl nitrile inhibitor of cathepsin C.
Biochem. Pharmacol., 164, 2019
|
|
1SZ7
 
 | | Crystal structure of Human Bet3 | | Descriptor: | PALMITIC ACID, Trafficking protein particle complex subunit 3 | | Authors: | Turnbull, A.P, Prinz, B, Holz, C, Behlke, J, Schultchen, J, Delbrueck, H, Niesen, F.H, Lang, C, Heinemann, U. | | Deposit date: | 2004-04-05 | | Release date: | 2005-01-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of palmitoylated BET3: insights into TRAPP complex assembly and membrane localization
Embo J., 24, 2005
|
|
7APJ
 
 | | Structure of autoinhibited Akt1 reveals mechanism of PIP3-mediated activation | | Descriptor: | NB41, RAC-alpha serine/threonine-protein kinase,Non-specific serine/threonine protein kinase,RAC-alpha serine/threonine-protein kinase | | Authors: | Truebestein, L, Hornegger, H, Leonard, T.A. | | Deposit date: | 2020-10-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of autoinhibited Akt1 reveals mechanism of PIP 3 -mediated activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ZQP
 
 | | Tail tip of siphophage T5 : open cone after interaction with bacterial receptor FhuA | | Descriptor: | Probable baseplate hub protein, Probable tape measure protein | | Authors: | Linares, R, Arnaud, C.A, Effantin, G, Darnault, C, Epalle, N, Boeri Erba, E, Schoehn, G, Breyton, C. | | Deposit date: | 2022-05-02 | | Release date: | 2023-02-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation.
Sci Adv, 9, 2023
|
|
5CZG
 
 | | Crystal Structure Analysis of hypothetical bromodomain Tb427.10.7420 from Trypanosoma brucei in complex with bromosporine | | Descriptor: | Bromosporine, Hypothetical Bromodomain, SODIUM ION, ... | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Amani, M, Hou, C.F.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-31 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Crystal Structure Analysis of hypothetical bromodomain from Trypanosoma brucei
to be published
|
|
3EKA
 
 | | Crystal structure of the complex of hyaluranidase trimer with ascorbic acid at 3.1 A resolution reveals the locations of three binding sites | | Descriptor: | ASCORBIC ACID, Hyaluronidase, phage associated | | Authors: | Mishra, P, Ethayathulla, A.S, Prem Kumar, R, Singh, N, Sharma, S, Kaur, P, Bhakuni, V, Singh, T.P. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Polysaccharide binding sites in hyaluronate lyase--crystal structures of native phage-encoded hyaluronate lyase and its complexes with ascorbic acid and lactose.
Febs J., 276, 2009
|
|
8A7X
 
 | | NaK C-DI F92A mutant soaked in Cs+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CESIUM ION, POTASSIUM ION, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2022-06-21 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors.
J.Mol.Biol., 435, 2023
|
|
3EL6
 
 | |
2IAM
 
 | | Structural basis for recognition of mutant self by a tumor-specific, MHC class II-restricted TCR | | Descriptor: | 15-mer peptide from Triosephosphate isomerase, CD4+ T cell receptor E8 alpha chain, CD4+ T cell receptor E8 beta chain, ... | | Authors: | Deng, L, Langley, R.J, Mariuzza, R.A. | | Deposit date: | 2006-09-08 | | Release date: | 2007-04-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor
Nat.Immunol., 8, 2007
|
|
7ZN4
 
 | | Tail tip of siphophage T5 : bent fibre after interaction with its bacterial receptor FhuA | | Descriptor: | Probable baseplate hub protein, Probable central straight fiber | | Authors: | Linares, R, Arnaud, C.A, Effantin, G, Darnault, C, Epalle, N, Boeri Erba, E, Schoehn, G, Breyton, C. | | Deposit date: | 2022-04-20 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation.
Sci Adv, 9, 2023
|
|
2W9R
 
 | | Structural basis of N-end rule substrate recognition in Escherichia coli by the ClpAP adaptor protein ClpS | | Descriptor: | ATP-DEPENDENT CLP PROTEASE ADAPTER PROTEIN CLPS, DNA PROTECTION DURING STARVATION PROTEIN | | Authors: | Schuenemann, V, Kralik, S.M, Albrecht, R, Spall, S.K, Truscott, K.N, Dougan, D.A, Zeth, K. | | Deposit date: | 2009-01-28 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of N-End Rule Substrate Recognition in Escherichia Coli by the Clpap Adaptor Protein Clps.
Embo Rep., 10, 2009
|
|
8ITM
 
 | | Cryo-EM structure of GIPR splice variant 2 (SV2) in complex with Gs protein | | Descriptor: | Gastric inhibitory polypeptide receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, F.H, Hang, K.N, Zhou, Q.T, Shao, L.J, Li, H, Li, W.Z, Lin, S, Dai, A.T, Cai, X.Q, Liu, Y.Y, Xu, Y.N, Feng, W.B, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-03-22 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Molecular basis of signal transduction mediated by the human GIPR splice variants.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7TBS
 
 | | Crystal Structure of the Glutaredoxin 2 from Francisella tularensis | | Descriptor: | CHLORIDE ION, Glutaredoxin 2, SULFATE ION | | Authors: | Kim, Y, Zhou, M, Grimshaw, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-22 | | Release date: | 2022-01-05 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of the Glutaredoxin 2 from Francisella tularensis
To Be Published
|
|
7ZLV
 
 | | Tail tip of siphophage T5 : central fibre protein pb4 | | Descriptor: | Probable central straight fiber | | Authors: | Linares, R, Arnaud, C.A, Effantin, G, Epalle, N, Boeri Erba, E, Schoehn, G, Breyton, C. | | Deposit date: | 2022-04-15 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation.
Sci Adv, 9, 2023
|
|
6W3L
 
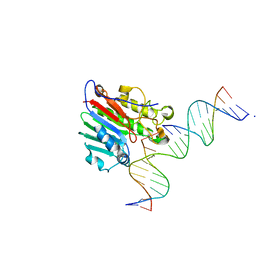 | | APE1 exonuclease substrate complex wild-type | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA-(apurinic or apyrimidinic site) lyase, ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2020-03-09 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
6P9G
 
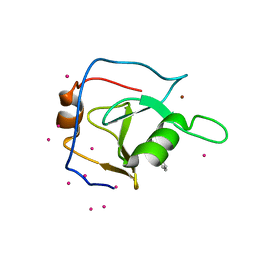 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with 2-(4-oxoquinazolin-3(4H)-yl)propanoic acid | | Descriptor: | (2R)-2-(4-oxoquinazolin-3(4H)-yl)propanoic acid, UNKNOWN ATOM OR ION, Ubiquitin carboxyl-terminal hydrolase 5, ... | | Authors: | Tempel, W, Mann, M.K, Harding, R.J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Schapira, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Small Molecule Antagonists of the USP5 Zinc Finger Ubiquitin-Binding Domain.
J.Med.Chem., 62, 2019
|
|
3EPP
 
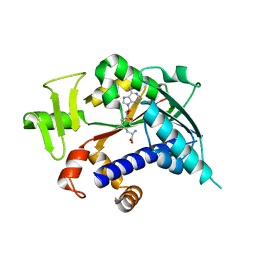 | | Crystal structure of mRNA cap guanine-N7 methyltransferase (RNMT) in complex with sinefungin | | Descriptor: | SINEFUNGIN, mRNA cap guanine-N7 methyltransferase | | Authors: | Amaya, M.F, Zeng, H, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Botchkarev, A, Min, J, Plotnikov, A.N, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-29 | | Release date: | 2008-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structure of mRNA cap guanine-N7 methyltransferase (RNMT) in complex with sinefungin.
To be Published
|
|
5KBT
 
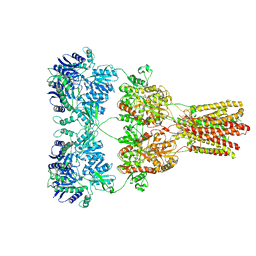 | | Cryo-EM structure of GluA2-1xSTZ complex at 6.4 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy.
Science, 353, 2016
|
|
8ITL
 
 | | Cryo-EM structure of GIPR splice variant 1 (SV1) in complex with Gs protein | | Descriptor: | Gastric inhibitory polypeptide receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, F.H, Hang, K.N, Zhou, Q.T, Shao, L.J, Li, H, Li, W.Z, Lin, S, Dai, A.T, Cai, X.Q, Liu, Y.Y, Xu, Y.N, Feng, W.B, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-03-22 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Molecular basis of signal transduction mediated by the human GIPR splice variants.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3ERW
 
 | | Crystal Structure of StoA from Bacillus subtilis | | Descriptor: | Sporulation thiol-disulfide oxidoreductase A | | Authors: | Crow, A, Liu, Y, Moller, M.C, Le Brun, N.E, Hederstedt, L. | | Deposit date: | 2008-10-03 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Functional Properties of Bacillus subtilis Endospore Biogenesis Factor StoA
J.Biol.Chem., 284, 2009
|
|
3E2K
 
 | | Crystal Structure of the KPC-2 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) | | Descriptor: | Beta-lactamase inhibitory protein, Carbapenemase | | Authors: | Hanes, M.S, Jude, K.M, Berger, J.M, Bonomo, R.A, Handel, T.M. | | Deposit date: | 2008-08-05 | | Release date: | 2009-08-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical characterization of the interaction between KPC-2 beta-lactamase and beta-lactamase inhibitor protein
Biochemistry, 48, 2009
|
|
3E32
 
 | | Protein farnesyltransferase complexed with FPP and ethylenediamine scaffold inhibitor 2 | | Descriptor: | FARNESYL DIPHOSPHATE, N-benzyl-N-(2-{(4-cyanophenyl)[(1-methyl-1H-imidazol-5-yl)methyl]amino}ethyl)-1-methyl-1H-imidazole-4-sulfonamide, Protein farnesyltransferase subunit beta, ... | | Authors: | Hast, M.A, Beese, L.S. | | Deposit date: | 2008-08-06 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for binding and selectivity of antimalarial and anticancer ethylenediamine inhibitors to protein farnesyltransferase.
Chem.Biol., 16, 2009
|
|
1TF9
 
 | | Streptomyces griseus aminopeptidase complexed with P-Iodo-L-Phenylalanine | | Descriptor: | Aminopeptidase, CALCIUM ION, IODO-PHENYLALANINE, ... | | Authors: | Reiland, V, Gilboa, R, Spungin-Bialik, A, Schomburg, D, Shoham, Y, Blumberg, S, Shoham, G. | | Deposit date: | 2004-05-27 | | Release date: | 2005-05-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Binding of inhibitory aromatic amino acids to Streptomyces griseus aminopeptidase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
