6VM3
 
 | | Full length Glycine receptor reconstituted in lipid nanodisc in Gly/IVM-conformation (State-3) | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Kumar, A, Basak, S, Chakrapani, S. | | Deposit date: | 2020-01-27 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Mechanisms of activation and desensitization of full-length glycine receptor in lipid nanodiscs.
Nat Commun, 11, 2020
|
|
6SF0
 
 | | Crystal Structure of Ancestral Flavin-containing monooxygenase (FMO) 2 in the presence of NADP+ | | Descriptor: | Ancestral Flavin-containing monooxygenase (FMO) 2, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Nicoll, C, Bailleul, G, Fiorentini, F, Mascotti, M.L, Fraaije, M, Mattevi, A. | | Deposit date: | 2019-07-30 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Ancestral-sequence reconstruction unveils the structural basis of function in mammalian FMOs.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7QYI
 
 | |
6FNJ
 
 | | Crystal Structure of Ephrin B4 (EphB4) Receptor Protein Kinase with an isomer of NVP-BHG712 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[(2-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]-~{N}-[3-(trifluoromethyl)phenyl]benzamide, Ephrin type-B receptor 4 | | Authors: | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Saxena, K, Schwalbe, H. | | Deposit date: | 2018-02-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.239 Å) | | Cite: | NVP-BHG712: Effects of Regioisomers on the Affinity and Selectivity toward the EPHrin Family.
ChemMedChem, 13, 2018
|
|
4S1V
 
 | |
7R5L
 
 | | Crystal structure of YTHDF2 with compound YLI_DC1_015 | | Descriptor: | 3,6-dimethyl-2~{H}-1,2,4-triazin-5-one, SULFATE ION, YTH domain-containing family protein 2 | | Authors: | Nachawati, R, Nai, F, Li, Y, Caflisch, A. | | Deposit date: | 2022-02-10 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fragment Ligands of the m 6 A-RNA Reader YTHDF2.
Acs Med.Chem.Lett., 13, 2022
|
|
5DIB
 
 | | 2.25 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) Y450L point mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-08-31 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2.25 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) Y450L point mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289
To Be Published
|
|
6LY8
 
 | | V/A-ATPase from Thermus thermophilus, the soluble domain, including V1, d, two EG stalks, and N-terminal domain of a-subunit. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Kishikawa, J, Nakanishi, A, Furuta, A, Kato, T, Namba, K, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2020-02-13 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanical inhibition of isolated V o from V/A-ATPase for proton conductance.
Elife, 9, 2020
|
|
6FTX
 
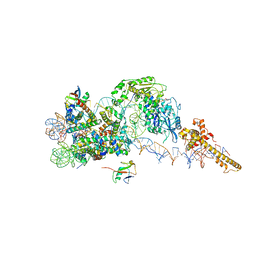 | | Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromatin-remodeling ATPase, ... | | Authors: | Sundaramoorthy, R, Owen-hughes, T, Norman, D.G, Hughes, A. | | Deposit date: | 2018-02-25 | | Release date: | 2018-08-08 | | Last modified: | 2018-10-17 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome.
Elife, 7, 2018
|
|
3OXE
 
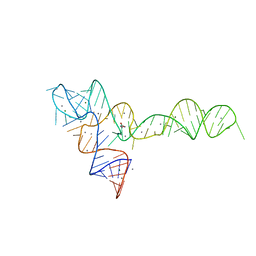 | | crystal structure of glycine riboswitch, Mn2+ soaked | | Descriptor: | GLYCINE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-21 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
6C1Z
 
 | |
5HKW
 
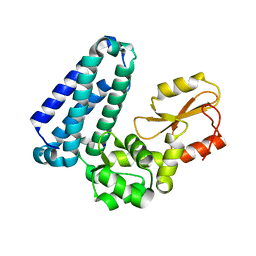 | | Crystal Structure of Apo c-Cbl TKBD Refined to 2.25 A Resolution | | Descriptor: | E3 ubiquitin-protein ligase CBL, SODIUM ION | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Apo c-Cbl TKBD Refined to 2.25 A Resolution
To be published
|
|
5D8N
 
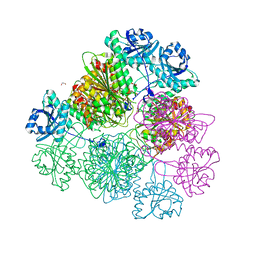 | | Tomato leucine aminopeptidase mutant - K354E | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | DuPrez, K.T, Scranton, M.A, Walling, L.L, Fan, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into chaperone-activity enhancement by a K354E mutation in tomato acidic leucine aminopeptidase.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6SEP
 
 | |
6SFG
 
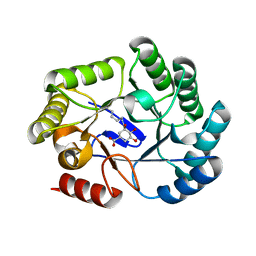 | | CRYSTAL STRUCTURE OF DHQ1 FROM SALMONELLA TYPHI COVALENTLY MODIFIED BY COMPOUND 9 | | Descriptor: | (1~{R},3~{S},4~{R},5~{R})-3-methyl-4,5-bis(hydroxyl)cyclohexane-1-carboxylic acid, (1~{S},3~{R},4~{S},5~{R})-3-methyl-3,4,5-tris(hydroxyl)cyclohexane-1-carboxylic Acid, 3-dehydroquinate dehydratase | | Authors: | Sanz-Gaitero, M, Lence, E, Maneiro, M, Thompson, R, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Self-Immolation of a Bacterial Dehydratase Enzyme by its Epoxide Product.
Chemistry, 26, 2020
|
|
6C3K
 
 | |
4NYA
 
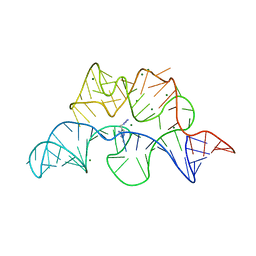 | | Crystal structure of the E. coli thiM riboswitch in complex with 5-(azidomethyl)-2-methylpyrimidin-4-amine | | Descriptor: | 5-(azidomethyl)-2-methylpyrimidin-4-amine, MAGNESIUM ION, thiM TPP riboswitch | | Authors: | Warner, K.D, Homan, P, Weeks, K.M, Smith, A.G, Abell, C, Ferre-D'Amare, A.R. | | Deposit date: | 2013-12-10 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Validating Fragment-Based Drug Discovery for Biological RNAs: Lead Fragments Bind and Remodel the TPP Riboswitch Specifically.
Chem.Biol., 21, 2014
|
|
5V3F
 
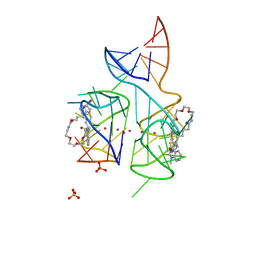 | | Co-crystal structure of the fluorogenic RNA Mango | | Descriptor: | 4-{[(2S)-3-{2,16-dioxo-20-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-6,9,12-trioxa-3,15-diazaicosan-1-yl}-2,3-dihydro-1,3-benzothiazol-2-yl]methyl}-1-methylquinolin-1-ium, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Trachman, R.J, Ferre-D'Amare, A.R. | | Deposit date: | 2017-03-07 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for high-affinity fluorophore binding and activation by RNA Mango.
Nat. Chem. Biol., 13, 2017
|
|
4FE1
 
 | | Improving the Accuracy of Macromolecular Structure Refinement at 7 A Resolution | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Fromme, R, Adams, P.D, Fromme, P, Levitt, M, Schroeder, G.F, Brunger, A.T. | | Deposit date: | 2012-05-29 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.9228 Å) | | Cite: | Improving the accuracy of macromolecular structure refinement at 7 A resolution.
Structure, 20, 2012
|
|
5YNX
 
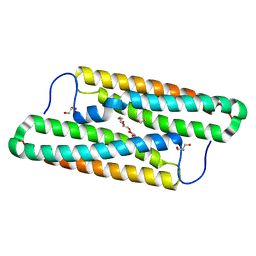 | | Structure of house dust mite allergen Der f 21 in PEG400 | | Descriptor: | Allergen Der f 21, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Ng, C.L, Chew, F.T, Pang, S.L, Ho, K.L, Teh, A.H, Waterman, J, Rambo, R, Mathavan, I, Beis, K, Say, Y.H. | | Deposit date: | 2017-10-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure and epitope analysis of house dust mite allergen Der f 21.
Sci Rep, 9, 2019
|
|
6SKW
 
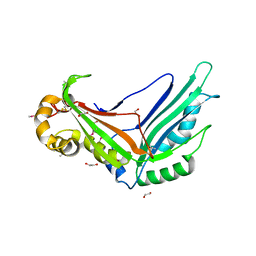 | | Crystal structure of the Legionella pneumophila type II secretion system substrate NttE | | Descriptor: | 1,2-ETHANEDIOL, NttE | | Authors: | Portlock, T.J, Rehman, S, Garnett, J.A. | | Deposit date: | 2019-08-16 | | Release date: | 2020-05-20 | | Last modified: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure, Dynamics and Cellular Insight Into Novel Substrates of theLegionella pneumophilaType II Secretion System.
Front Mol Biosci, 7, 2020
|
|
3JUT
 
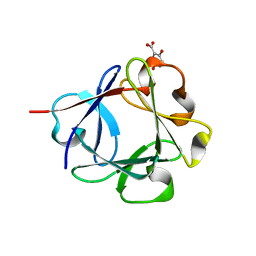 | | Acidic Fibroblast Growth Factor (FGF-1) complexed with gentisic acid | | Descriptor: | 2,5-dihydroxybenzoic acid, Heparin-binding growth factor 1 | | Authors: | Fernandez, I.S, Gimenez-Gallego, G, Romero, A. | | Deposit date: | 2009-09-15 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Gentisic acid, a compound associated with plant defense and a metabolite of aspirin, heads a new class of in vivo fibroblast growth factor inhibitors.
J.Biol.Chem., 285, 2010
|
|
6G4T
 
 | |
4ZOB
 
 | | Crystal Structure of beta-glucosidase from Listeria innocua in complex with gluconolactone | | Descriptor: | D-glucono-1,5-lactone, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
6SNL
 
 | | (R)-selective amine transaminase from Exophiala sideris | | Descriptor: | CHLORIDE ION, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Telzerow, A, Hakansson, M, Steiner, K. | | Deposit date: | 2019-08-26 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.129 Å) | | Cite: | Expanding the Toolbox of R-Selective Amine Transaminases by Identification and Characterization of New Members.
Chembiochem, 22, 2021
|
|
