5N83
 
 | |
8DT0
 
 | |
6WD4
 
 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure II-B2) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
7ONP
 
 | | Wild type carbonic anhydrase II with bound IrCp* complex to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 4-[2-(9-chloranyl-2',3',4',5',6'-pentamethyl-4-nitro-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, ACETATE ION, Carbonic anhydrase 2, ... | | Authors: | Stein, A, Dongping, C, Cotelle, Y, Rebelein, J.G, Ward, T.R. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.408 Å) | | Cite: | A Dual Anchoring Strategy for the Directed Evolution of Improved Artificial Transfer Hydrogenases Based on Carbonic Anhydrase.
Acs Cent.Sci., 7, 2021
|
|
6WDZ
 
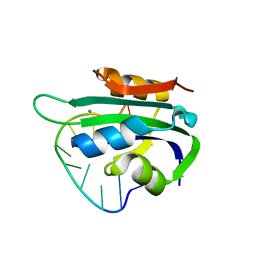 | | Porcine circovirus 2 Rep domain complexed with a single-stranded DNA 10-mer comprising the cleavage site | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent helicase Rep, DNA (5'-D(*TP*AP*GP*TP*AP*TP*TP*AP*CP*C)-3'), ... | | Authors: | Litzau, L.A, Tompkins, K, Shi, K, Nelson, A, Evans III, R.L, Gordon, W.R. | | Deposit date: | 2020-04-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular underpinnings of ssDNA specificity by Rep HUH-endonucleases and implications for HUH-tag multiplexing and engineering.
Nucleic Acids Res., 49, 2021
|
|
7S57
 
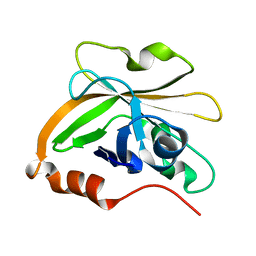 | | Structure of Sortase A from Streptococcus pyogenes with the b7-b8 loop sequence of Enterococcus faecalis Sortase A | | Descriptor: | Class A sortase, sortase A chimera | | Authors: | Svendsen, J.E, Johnson, D.A, Gao, M, Antos, J.M, Amacher, J.F. | | Deposit date: | 2021-09-09 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical analyses of selectivity determinants in chimeric Streptococcus Class A sortase enzymes.
Protein Sci., 31, 2022
|
|
8DUH
 
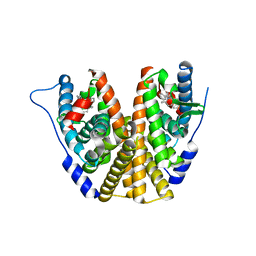 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with (6'-hydroxy-1'-(4-(2-((R)-2-methylpyrrolidin-1-yl)ethoxy)phenyl)-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl)(phenyl)methanone | | Descriptor: | Estrogen receptor, [(1'R)-6'-hydroxy-1'-(4-{2-[(2R)-2-methylpyrrolidin-1-yl]ethoxy}phenyl)-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl](phenyl)methanone | | Authors: | Hancock, G.R, Young, K.S, Hosfield, D.J, Joiner, C, Sullivan, E.A, Yildz, Y, Laine, M, Greene, G.L, Fanning, S.W. | | Deposit date: | 2022-07-27 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unconventional isoquinoline-based SERMs elicit fulvestrant-like transcriptional programs in ER+ breast cancer cells.
NPJ Breast Cancer, 8, 2022
|
|
7S53
 
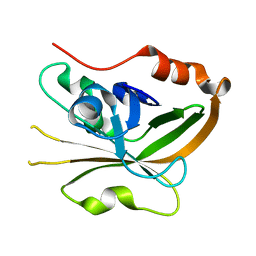 | | Structure of Sortase A from Streptococcus pyogenes with the b7-b8 loop sequence from Listeria monocytogenes Sortase A | | Descriptor: | Class A sortase, sortase A chimera | | Authors: | Johnson, D.A, Svendsen, J.E, Antos, J.M, Amacher, J.F. | | Deposit date: | 2021-09-09 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and biochemical analyses of selectivity determinants in chimeric Streptococcus Class A sortase enzymes.
Protein Sci., 31, 2022
|
|
6WDA
 
 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure III-C) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
6WE1
 
 | | Wheat dwarf virus Rep domain complexed with a single-stranded DNA 8-mer comprising the cleavage site | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*TP*TP*AP*C)-3'), MANGANESE (II) ION, Replication-associated protein | | Authors: | Tompkins, K, Litzau, L.A, Pornschloegl, L, Nelson, A.T, Evans III, R.L, Gordon, W.R. | | Deposit date: | 2020-04-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.612 Å) | | Cite: | Molecular underpinnings of ssDNA specificity by Rep HUH-endonucleases and implications for HUH-tag multiplexing and engineering.
Nucleic Acids Res., 49, 2021
|
|
8DUS
 
 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with (1'-(4-(2-(ethylamino)ethoxy)phenyl)-6'-hydroxy-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl)(phenyl)methanone | | Descriptor: | Estrogen receptor, [(1'R)-1'-{4-[2-(ethylamino)ethoxy]phenyl}-6'-hydroxy-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl](phenyl)methanone | | Authors: | Hancock, G.R, Young, K.S, Hosfield, D.J, Joiner, C, Sullivan, E.A, Yildz, Y, Laine, M, Greene, G.L, Fanning, S.W. | | Deposit date: | 2022-07-27 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unconventional isoquinoline-based SERMs elicit fulvestrant-like transcriptional programs in ER+ breast cancer cells.
NPJ Breast Cancer, 8, 2022
|
|
7BNS
 
 | | VDR complex with BXL-62 | | Descriptor: | 1,25-Dihydroxy-16-ene-20-cyclopropyl-vitamin D3, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N, Belorusova, A.Y. | | Deposit date: | 2021-01-22 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Vitamin D Analogs Bearing C-20 Modifications Stabilize the Agonistic Conformation of Non-Responsive Vitamin D Receptor Variants.
Int J Mol Sci, 23, 2022
|
|
8UOA
 
 | | Structure of the synaptic vesicle protein 2A Luminal domain in complex with a nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody, Synaptic vesicle glycoprotein 2A, ... | | Authors: | Mittal, A, Martin, M.F, Levin, E, Adams, C, Yang, M, Ledecq, M, Horanyi, P.S, Coleman, J.A. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of synaptic vesicle protein 2A and 2B bound to anticonvulsants.
Nat.Struct.Mol.Biol., 2024
|
|
7RXT
 
 | | Crystal of BRD4(D1) with 2-[(3R)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine | | Descriptor: | 2-[(3R)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine, Bromodomain-containing protein 4 | | Authors: | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
8UO8
 
 | | Structure of synaptic vesicle protein 2B with padsevonil | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, (4R)-4-(2-chloro-2,2-difluoroethyl)-1-{[(4R)-2-(methoxymethyl)-6-(trifluoromethyl)imidazo[2,1-b][1,3,4]thiadiazol-5-yl]methyl}pyrrolidin-2-one, 1,2-DIDECANOYL-SN-GLYCERO-3-[PHOSPHO-L-SERINE], ... | | Authors: | Martin, M.F, Mittal, A, Levin, E, Adams, C, Yang, M, Ledecq, M, Horanyi, P.S, Coleman, J.A. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of synaptic vesicle protein 2A and 2B bound to anticonvulsants.
Nat.Struct.Mol.Biol., 2024
|
|
5KU8
 
 | | Crystal structure of CK2 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Ferguson, A.D, Dowling, J. | | Deposit date: | 2016-07-13 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of CK2
Not Published
|
|
6ZAC
 
 | | PI3K Delta in complex with [(dimethylamino)methyldihydrobenzoxazin2methoxypyridinyl]methanesulfonamide | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ~{N}-[5-[6-[(dimethylamino)methyl]-2,3-dihydro-1,4-benzoxazin-4-yl]-2-methoxy-pyridin-3-yl]methanesulfonamide | | Authors: | Convery, M.A, Hardy, C.J, Spencer, J.A, Rowland, P. | | Deposit date: | 2020-06-05 | | Release date: | 2020-07-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design and Development of a Macrocyclic Series Targeting Phosphoinositide 3-Kinase delta.
Acs Med.Chem.Lett., 11, 2020
|
|
8DUG
 
 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with (6'-hydroxy-1'-(4-(2-((S)-3-methylpyrrolidin-1-yl)ethoxy)phenyl)-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl)(phenyl)methanone | | Descriptor: | Estrogen receptor, [(1'R)-6'-hydroxy-1'-(4-{2-[(3R)-3-methylpyrrolidin-1-yl]ethoxy}phenyl)-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl](phenyl)methanone | | Authors: | Hancock, G.R, Young, K.S, Hosfield, D.J, Joiner, C, Sullivan, E.A, Yildz, Y, Laine, M, Greene, G.L, Fanning, S.W. | | Deposit date: | 2022-07-27 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unconventional isoquinoline-based SERMs elicit fulvestrant-like transcriptional programs in ER+ breast cancer cells.
NPJ Breast Cancer, 8, 2022
|
|
6YSQ
 
 | | The hC4Nb8 complement inhibitory nanobody in complex with C4b | | Descriptor: | Complement C4 beta chain, Complement C4 gamma chain, Complement C4-A alpha chain, ... | | Authors: | Zarantonello, A, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | An Ultrahigh-Affinity Complement C4b-Specific Nanobody Inhibits In Vivo Assembly of the Classical Pathway Proconvertase.
J Immunol., 205, 2020
|
|
6YTA
 
 | | CLK1 bound with imidazopyridazine (Cpd 1) | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-{6-[(CYCLOPROPYLMETHYL)AMINO]IMIDAZO[1,2-B]PYRIDAZIN-3-YL}PHENYL)ETHANONE, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6WEK
 
 | | Structure of cGMP-bound WT TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CYCLIC GUANOSINE MONOPHOSPHATE, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8E4R
 
 | |
6YTG
 
 | | CLK1 bound with beta-carboline KH-CARB13 (Cpd 3) | | Descriptor: | (4~{S})-7,8-bis(chloranyl)-9-methyl-1-oxidanylidene-spiro[2,4-dihydropyrido[3,4-b]indole-3,4'-piperidine]-4-carbonitrile, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Chaikuad, A, Huber, K, Bracher, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
8VQ4
 
 | | CDK2-CyclinE1 in complex with allosteric inhibitor I-125A. | | Descriptor: | (8R)-6-(1-benzyl-1H-pyrazole-4-carbonyl)-N-[(2S,3R)-3-(2-cyclohexylethoxy)-1-(methylamino)-1-oxobutan-2-yl]-2-[(1S)-2,2-dimethylcyclopropane-1-carbonyl]-2,6-diazaspiro[3.4]octane-8-carboxamide, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Hirschi, M, Johnson, E, Zhang, Y, Liu, Z, Brodsky, O, Won, S.J, Nagata, A, Petroski, M.D, Majmudar, J.D, Niessen, S, VanArsdale, T, Gilbert, A.M, Hayward, M.M, Stewart, A.E, Nager, A.R, Melillo, B, Cravatt, B. | | Deposit date: | 2024-01-17 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Expanding the ligandable proteome by paralog hopping with covalent probes.
Biorxiv, 2024
|
|
6YU7
 
 | | Crystal structure of MhsT in complex with L-tyrosine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, SODIUM ION, Sodium-dependent transporter, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
