5V0Y
 
 | | Solution structure of arenicin-3. | | Descriptor: | arenicin-3 | | Authors: | Edwards, I.A, Mobli, M. | | Deposit date: | 2017-02-28 | | Release date: | 2018-08-08 | | Last modified: | 2020-06-10 | | Method: | SOLUTION NMR | | Cite: | An amphipathic peptide with antibiotic activity against multidrug-resistant Gram-negative bacteria
Nat Commun, 2020
|
|
7ZLG
 
 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in complex with acceptor peptide and bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mukherjee, S, Mao, R, Irobalieva, R, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
2W2B
 
 | | Crystal Structure of single point mutant Tyr20Phe p-coumaric Acid Decarboxylase from Lactobacillus plantarum: structural insights into the active site and decarboxylation catalytic mechanism | | Descriptor: | ACETATE ION, ISOPROPYL ALCOHOL, P-COUMARIC ACID DECARBOXYLASE | | Authors: | Rodriguez, H, Angulo, I, de las Rivas, B, Campillo, N, Paez, J.A, Munoz, R, Mancheno, J.M. | | Deposit date: | 2008-10-27 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | P-Coumaric Acid Decarboxylase from Lactobacillus Plantarum: Structural Insights Into the Active Site and Decarboxylation Catalytic Mechanism.
Proteins, 78, 2010
|
|
7ZLJ
 
 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in ternary complex with Dol25-P-C-Man and acceptor peptide, bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mao, R, Mukherjee, S, Boilevin, J, Irobalieva, R, Darbre, T, Reymond, J.L, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
1TBR
 
 | |
6PI0
 
 | |
7ZLI
 
 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in complex with Dol25-P-Man and bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mukherjee, S, Boilevin, J, Irobalieva, R, Darbre, T, Reymond, J.L, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
3EE2
 
 | |
8IOD
 
 | | Cryo-EM structure of the PG-901-bound human melanocortin receptor 5 (MC5R)-Gs complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1,HiBiT, ... | | Authors: | Feng, W.B, Zhou, Q.T, Chen, X.Y, Dai, A.T, Cai, X.Q, Liu, X, Zhao, F.H, Chen, Y, Ye, C.Y, Xu, Y.N, Cong, Z.T, Li, H, Lin, S, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-03-10 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural insights into ligand recognition and subtype selectivity of the human melanocortin-3 and melanocortin-5 receptors.
Cell Discov, 9, 2023
|
|
8INR
 
 | | Cryo-EM structure of the alpha-MSH-bound human melanocortin receptor 5 (MC5R)-Gs complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1,HiBiT, ... | | Authors: | Feng, W.B, Zhou, Q.T, Chen, X.Y, Dai, A.T, Cai, X.Q, Liu, X, Zhao, F.H, Chen, Y, Ye, C.Y, Xu, Y.N, Cong, Z.T, Li, H, Lin, S, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-03-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural insights into ligand recognition and subtype selectivity of the human melanocortin-3 and melanocortin-5 receptors.
Cell Discov, 9, 2023
|
|
1STA
 
 | | ACCOMMODATION OF INSERTION MUTATIONS ON THE SURFACE AND IN THE INTERIOR OF STAPHYLOCOCCAL NUCLEASE | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Keefe, L.J, Quirk, S, Gittis, A, Lattman, E.E. | | Deposit date: | 1994-01-17 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Accommodation of insertion mutations on the surface and in the interior of staphylococcal nuclease.
Protein Sci., 3, 1994
|
|
6PNL
 
 | | Structure of Epimerase Mth375 from the thermophilic pseudomurein-containing methanogen Methanothermobacter thermautotrophicus | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of a UDP-GALE 4-epimerase (Mth375) from the thermophilic pseudomurein-containing methanogen Methanothermobacter thermautotrophicus
To Be Published
|
|
8IOC
 
 | | Cryo-EM structure of the gamma-MSH-bound human melanocortin receptor 3 (MC3R)-Gs complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1,HiBiT, ... | | Authors: | Feng, W.B, Zhou, Q.T, Chen, X.Y, Dai, A.T, Cai, X.Q, Liu, X, Zhao, F.H, Chen, Y, Ye, C.Y, Xu, Y.N, Cong, Z.T, Li, H, Lin, S. | | Deposit date: | 2023-03-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural insights into ligand recognition and subtype selectivity of the human melanocortin-3 and melanocortin-5 receptors.
Cell Discov, 9, 2023
|
|
4R0Z
 
 | | A conserved phosphorylation switch controls the interaction between cadherin and beta-catenin in vitro and in vivo | | Descriptor: | FORMIC ACID, Protein humpback-2 | | Authors: | Choi, H.-J, Loveless, T, Lynch, A, Bang, I, Hardin, J, Weis, W.I. | | Deposit date: | 2014-08-03 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | A Conserved Phosphorylation Switch Controls the Interaction between Cadherin and beta-Catenin In Vitro and In Vivo
Dev.Cell, 33, 2015
|
|
2VWB
 
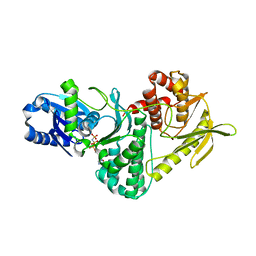 | | Structure of the archaeal Kae1-Bud32 fusion protein MJ1130: a model for the eukaryotic EKC-KEOPS subcomplex involved in transcription and telomere homeostasis. | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PUTATIVE O-SIALOGLYCOPROTEIN ENDOPEPTIDASE | | Authors: | Hecker, A, Lopreiato, R, Graille, M, Collinet, B, Forterre, P, Domenico, L, van Tilbeurgh, H. | | Deposit date: | 2008-06-20 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of the Archaeal Kae1/Bud32 Fusion Protein Mj1130: A Model for the Eukaryotic Ekc/Keops Subcomplex
Embo J., 27, 2008
|
|
3WY8
 
 | | Crystal Structure of Protease Anisep from Arthrobacter Nicotinovorans | | Descriptor: | Serine protease | | Authors: | Sone, T, Haraguchi, Y, Kuwahara, A, Ose, T, Takano, M, Abe, A, Tanaka, M, Tanaka, I, Asano, K. | | Deposit date: | 2014-08-20 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization reveals the keratinolytic activity of an arthrobacter nicotinovorans protease.
Protein Pept.Lett., 22, 2015
|
|
2I7N
 
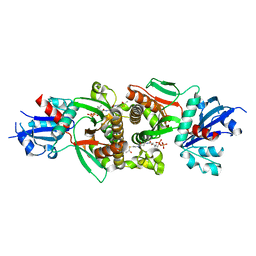 | | Crystal structure of human PANK1 alpha: the catalytic core domain in complex with AcCoA | | Descriptor: | ACETYL COENZYME *A, Pantothenate kinase 1 | | Authors: | Hong, B.S, Wang, L, Tempel, W, Loppnau, P, Allali-Hassani, A, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H.W. | | Deposit date: | 2006-08-31 | | Release date: | 2006-12-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of human pantothenate kinases. Insights into allosteric regulation and mutations linked to a neurodegeneration disorder.
J.Biol.Chem., 282, 2007
|
|
1SYV
 
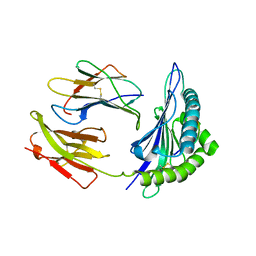 | | HLA-B*4405 complexed to the dominant self ligand EEFGRAYGF | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, major histocompatibility complex, ... | | Authors: | Zernich, D, Purcell, A.W, Macdonald, W.A, Kjer-Nielsen, L, Ely, L.K, Laham, N, Crockford, T, Mifsud, N.A, Tait, B.D, Holdsworth, R, Brooks, A.G, Bottomley, S.P, Beddoe, T, Peh, C.A, Rossjohn, J, McCluskey, J. | | Deposit date: | 2004-04-02 | | Release date: | 2004-10-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Natural HLA class I polymorphism controls the pathway of antigen presentation and susceptibility to viral evasion
J.Exp.Med., 200, 2004
|
|
4R4Q
 
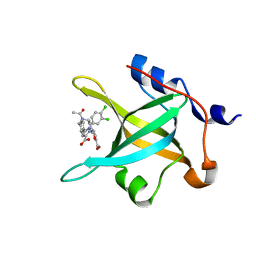 | | Crystal structure of RPA70N in complex with 5-(3-((N-(4-(5-carboxyfuran-2-yl)benzyl)acetamido)methyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | Descriptor: | 5-[4-({acetyl[4-(5-carboxyfuran-2-yl)benzyl]amino}methyl)phenyl]-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
3EKK
 
 | | Insulin receptor kinase complexed with an inhibitor | | Descriptor: | 2-[(2-{[1-(N,N-dimethylglycyl)-5-methoxy-1H-indol-6-yl]amino}-7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]-6-fluoro-N-methylbenzamide, Insulin receptor | | Authors: | Chamberlain, S, Atkins, C, Deanda, F, Dumble, M, Gerding, R, Groy, A, Korenchuk, S, Kumar, R, Lei, H, Mook, R, Moorthy, G, Redman, A, Rowland, J, Sabbatini, P, Shewchuk, L. | | Deposit date: | 2008-09-19 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of 4,6-bis-anilino-1H-pyrrolo[2,3-d]pyrimidines: Potent inhibitors of the IGF-1R receptor tyrosine kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2W4X
 
 | | BtGH84 in complex with STZ | | Descriptor: | CALCIUM ION, GLYCEROL, O-GLCNACASE BT_4395, ... | | Authors: | He, Y, Bubb, A, Martinez-Fleites, C, Davies, G.J. | | Deposit date: | 2008-12-02 | | Release date: | 2009-02-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Insight Into the Mechanism of Streptozotocin Inhibition of O-Glcnacase.
Carbohydr.Res., 344, 2009
|
|
4W6Y
 
 | | Co-complex structure of the lectin domain of F18 fimbrial adhesin FedF with inhibitory nanobody NbFedF9 | | Descriptor: | F18 fimbrial adhesin AC, Nanobody NbFedF9, SULFATE ION | | Authors: | Moonens, K, De Kerpel, M, Coddens, A, Cox, E, Pardon, E, Remaut, H, De Greve, H. | | Deposit date: | 2014-08-21 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Nanobody Mediated Inhibition of Attachment of F18 Fimbriae Expressing Escherichia coli.
Plos One, 9, 2014
|
|
3ELJ
 
 | | Jnk1 complexed with a bis-anilino-pyrrolopyrimidine inhibitor. | | Descriptor: | 2-fluoro-6-{[2-({2-methoxy-4-[(methylsulfonyl)methyl]phenyl}amino)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]amino}benzamide, Mitogen-activated protein kinase 8 | | Authors: | Chamberlain, S, Atkins, C, Deanda, F, Dumble, M, Gerding, R, Groy, A, Korenchuk, S, Kumar, R, Lei, H, Mook, R, Moorthy, G, Redman, A, Rowland, J, Shewchuk, L, Vicentini, G, Mosley, J. | | Deposit date: | 2008-09-22 | | Release date: | 2008-12-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Optimization of 4,6-bis-anilino-1H-pyrrolo[2,3-d]pyrimidine IGF-1R tyrosine kinase inhibitors towards JNK selectivity.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3EE8
 
 | |
1T4A
 
 | | Structure of B. Subtilis PurS C2 Crystal Form | | Descriptor: | PurS | | Authors: | Anand, R, Hoskins, A.A, Bennett, E.M, Sintchak, M.D, Stubbe, J, Ealick, S.E. | | Deposit date: | 2004-04-28 | | Release date: | 2004-09-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A model for the Bacillus subtilis formylglycinamide ribonucleotide amidotransferase multiprotein complex.
Biochemistry, 43, 2004
|
|
